- EN - English
- CN - 中文
Purification and Proteomic Analysis of Alphavirus Particles from Sindbis Virus Grown in Mammalian and Insect Cells
哺乳动物和昆虫细胞中的辛德比斯病毒的α病毒颗粒的纯化和蛋白质组分析
(*contributed equally to this work) 发布: 2019年05月20日第9卷第10期 DOI: 10.21769/BioProtoc.3239 浏览次数: 7514
评审: David PaulJinping ZhaoAnonymous reviewer(s)

相关实验方案

诱导型HIV-1库削减检测(HIVRRA):用于评估外周血单个核细胞中HIV-1潜伏库清除策略毒性与效力的快速敏感方法
Jade Jansen [...] Neeltje A. Kootstra
2025年07月20日 2460 阅读
Abstract
Current mass spectrometry (MS) methods and new instrumentation now allow for more accurate identification of proteins in low abundance than previous protein fractionation and identification methods. It was of interest if this method could serve to define the virus proteome of a membrane-containing virus. To evaluate the efficacy of mass spec to determine the proteome of medically important viruses, Sindbis virus (SINV), the prototypical alphavirus was chosen for evaluation. This model system was chosen specifically because the alphaviruses contain members which are human pathogens, this virus is well defined biochemically and structurally, and grows to high titers in both vertebrate and non-vertebrate host cells. The SINV proteome was investigated using this method to determine if host proteins are specifically packaged into infectious virions. It was also of interest if the SINV proteome, when grown in multiple host cells representing vertebrate and mosquito hosts, incorporated specific host proteins from all hosts. Observation of recurrent or distinctive proteins in the virus proteome aided in the determination of proteins incorporated into the virion as opposed to those bound to the particle exterior. Mass spectrometry analysis identified the total protein content of purified virions within limits of detection. The most significant finding was that in addition to the host proteins, SINV non-structural protein 2 (nsP2) was detected within virions grown in all host cells examined. This analysis identified host factors not previously associated with alphavirus entry, replication, or egress, identifying at least one host factor integrally involved in alphavirus replication. Key to the success of this analysis is the method of virus purification which must deliver measurably infectious virus free of high levels of contaminants. For SINV and other members of the alphavirus family, this is accomplished by isopycnic centrifugation through potassium tartrate, followed by a high salt wash.
Keywords: Sindbis virus (辛德比斯病毒)Background
The terms proteome and proteomics were coined by Marc Wilkins to describe the systematic evaluation of proteins in a model system using a detailed study of structure, function and regulation of its biology including aberrations which lead to disease (Wilkins et al., 1996 and 2009). However, virus proteomes have been under investigation long before the field of proteomics evolved in an attempt to understand the mechanisms of virus-host interactions in vitro and to evaluate virus pathogenesis of the animal host. To this end it was of interest to evaluate the virus proteome of a model system in a family of viruses which contained medically significant pathogens. Sindbis virus was chosen for this investigation because it is a member of the Alphavirus genus, Family Togaviridae, which contains a significant number of human pathogens of medical importance, it is structurally stable, grows to high titers, is well described in the literature and can be grown in vertebrate and non-vertebrate host cells. Sindbis virus has been the subject of many studies because it is a relatively simple membrane-containing +RNA icosahedral virus (Figure 1). The viral 42S genome is infectious and serves as the template for a 26S subgenomic RNA which encodes the structural genes, organized in the sequence C-PE2(E3/E2)-6K(TF)-E1 3’UTR-polyA (Figure 2). The genomic strand encodes the non-structural proteins in the order 5’UTR-nsP1-nsP2-nsP3-nsP4 (Strauss, J. H. and Strauss, E. G., 1994). The process of virus assembly has marked differences when occurring in vertebrate or invertebrate cells (Brown, 1986). In vertebrate cells the nucleocapsid is pre-assembled in the cytoplasm with the genomic RNA prior to association with the viral protein modified plasma membrane and virus budding from the plasma membrane. In invertebrate cells the virus nucleocapsid and fully matured virus are assembled in endosomes termed “virus factories” prior to fusion of the virus-containing endosomes with the plasma membrane, releasing infectious particles. Depending on the virus’ host cells origin, the glycosylation patterns of the glycoproteins will be that specified by the cellular biochemistry and the specific lipid bilayer will be that of the host cell. The virus particles released from either host can be equally infectious and if the SVHR strain of Sindbis virus is used close to 100% viability is achieved (Vancini et al., 2013). This is an important detail because virus infectivity requires all of its components to be in their metastable conformation and native physical state to be functional that is, infectious (Hernandez et al., 2014). Thus, not all virus particles will be amenable to this method of analysis because the particle structure must be 1) of regular stoichiometry 2) capable of rigorous purification without the loss of infectivity and 3) express a very low particle to PFU ratio. These factors are important to be able to discern any protein contaminants from proteins that are carried within the virus particle. Because of the symmetry of the virus particle and the stoichiometry, the protein concentration can be used to calculate the number of physical particles from the number of infectious particles using the plaque assay to determine infectious particle titer. This specific analysis cannot be applied to non-symmetrical viruses.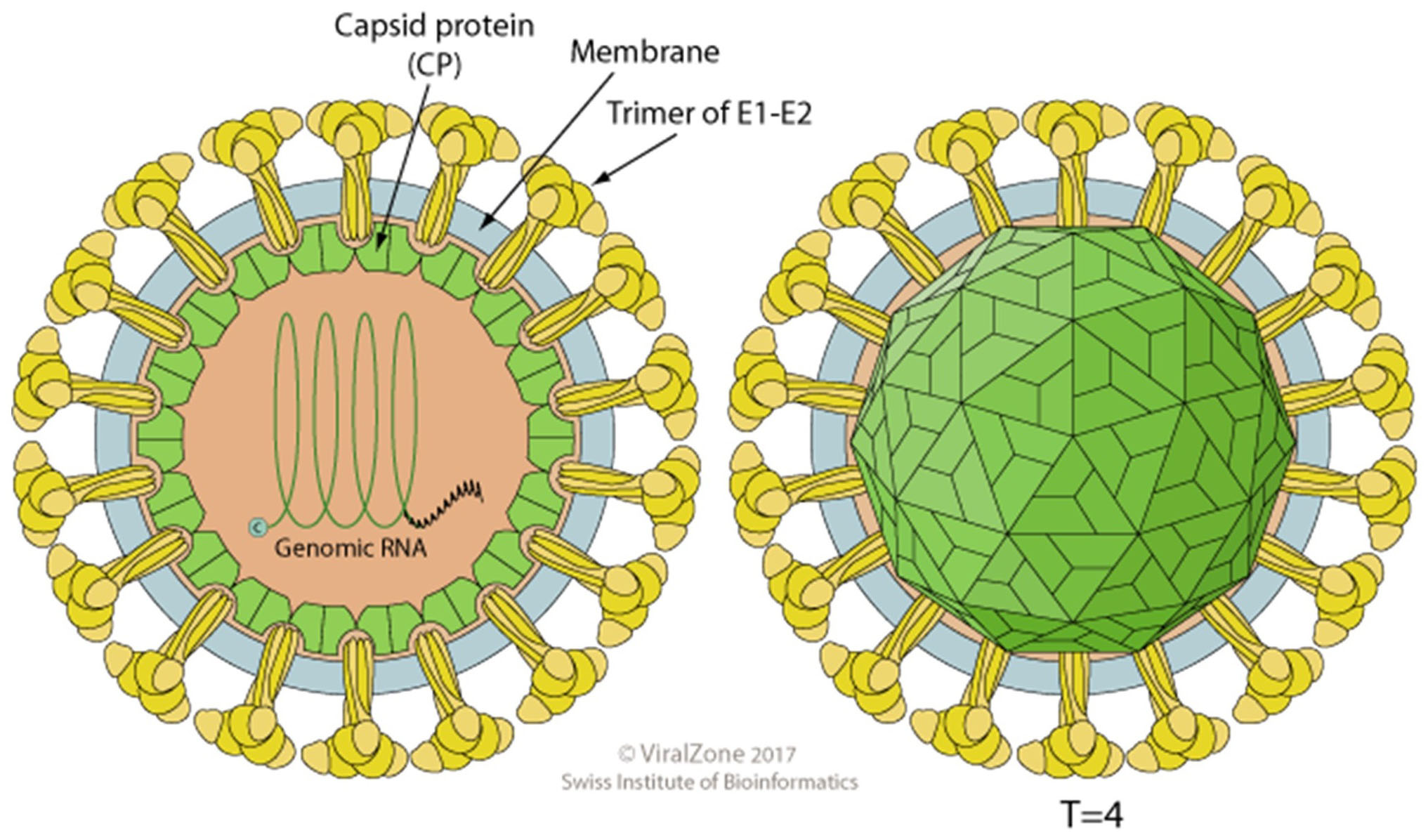
Figure 1. A cartoon of an alphavirus. The cartoon depicts an enveloped, spherical, icosahedral, 65-70 nm in diameter, capsid with a T = 4 icosahedral symmetry made of 240 monomers 1:1:1 stoichiometric ratio of E1:E2:C. The envelope contains 80 spikes, each spike is a trimer of E1/E2 protein dimers. Reprinted with permission from ViralZone, SBI Swiss Institute of Bioinformatics. Figures from ViralZone, with permission.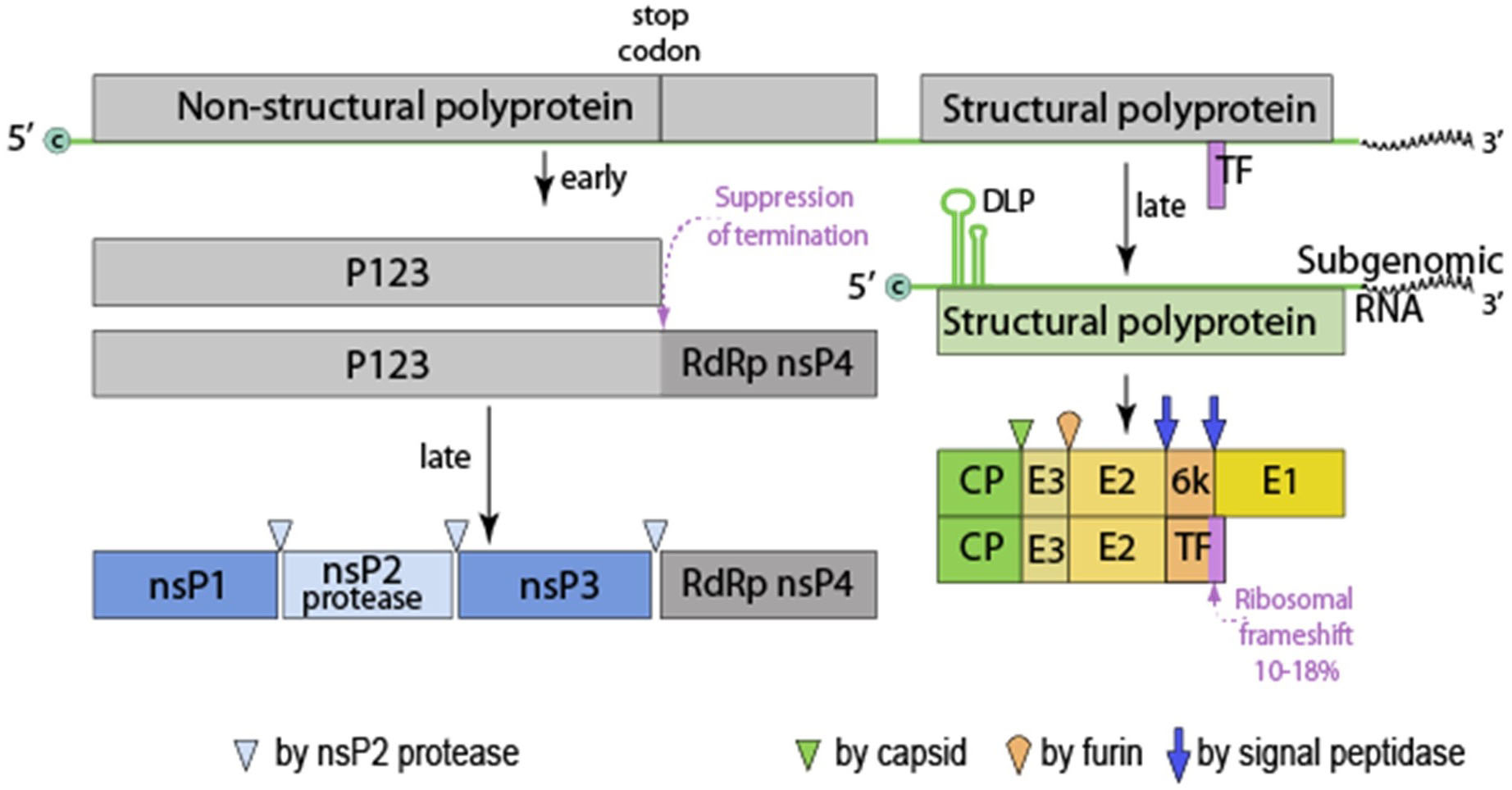
Figure 2. Alphavirus genomic organization. This genome is monopartite, linear, ssRNA(+) genome of 11-12 kb. The genome is capped (c) and polyadenylated. P123, precursor to nsP1, 2 and 3. Virus proteins are matured by both host and viral proteases. RdRp, RNA dependent RNA polymerase, TF, transframe protein (made from ribosomal frame shift in 6k protein) and DLP (downstream loop), stable RNA structure in the coding sequence of the 5’ region of the subgenomic RNA. Figures from ViralZone, with permission.
Stock Virus growth in BHK cells or C7-10 cells
The heat resistant SINV (SVHR) strain was used in this study. This strain was isolated by Burge and Pfefferkorn in 1966 by collecting virus that was resistant to heating to 54 °C. The choice of virus strain is important because this strain produces high titers (1010 PFU/ml) and low particle/PFU (~1 particle/PFU), ratios of highly infectious and physically stable virus. BHK and C7-10 (Aedes albopictus) cells were obtained from internal collections and are the favored cells to grow SVHR. Virus was harvested from 10 T-75 flasks (Corning) which produces enough virus to form a large visible band in a 30 ml potassium tartrate gradient and sufficient material for the mass spec analysis. Cells were infected at an MOI of 10 PFU/ml, for Sindbis virus infections and allowed to replicate for a single cycle and harvested at 18 h post infection to ensure that no cell lysis took place. A single cycle of SINV growth from mammalian cells is ~12 h and from C7-10 cells is about 24 h. The supernatants were clarified by low speed centrifugation (Sorvall RC-5B Super speed centrifuge at 2,000 rpm, 700 x g). Twenty microliter of the resulting virus supernatant was loaded onto a 15-35% linear potassium tartrate gradient and twice purified by isopycnic ultracentrifugation (Beckman SW-28 rotor, 18 h at 10,000 x g). The resulting band of purified virus was collected and washed twice by pelleting the virus in 5 ml 1x PBS in an SW-40 Beckman-40 rotor at 45,000 rpm (12,000 x g) for 30 min and collecting the pellet.
Virus titration by Plaque assay
The assay of virus titer by plaque formation, “plaque assay” is the most accurate method for measuring of the amount of infectious virus. This assay is used to determine the titer, in plaque-forming units (PFU) per ml, of virus by infecting a standardized monolayer of cells with a known volume of a known dilution of a virus-containing solution. The infection is contained under agarose which only allows diffusion of the virus to adjacent cells. Virus from a single initially infected cell infects adjacent cells producing a “plaque” or clearing (formed by lysed cells) localized to the original site of infection by an overlay of 1% agarose in 1x EMEM. Plaques of SVHR are visible to the naked eye after neutral red staining within 2-3 days of incubation at 37 °C. Begin with an estimate of what the titer could be, if the titer is estimated to be around 108 PFU/ml, a flask infected with a dilution of 10-6 would show 20-200 plaques; in this case, infecting flasks with dilutions of 10-5, 10-6, and 10-7 PFU/ml should give adequate data to make a relatively accurate calculation. If the titer of the virus is completely unknown, it may be necessary to infect flasks or plates with a wide range of dilutions (10-1 to 10-8). The number and quality of the plaques seen in a given assay can be influenced by a number of factors, including the pH and/or temperature of media, dilution buffer, agarose overlay, or the condition of the cell monolayer. Due to the sensitivity of this assay, it is important to include both positive and negative controls within each assay. The negative control is a flask that is inoculated with diluent only, and the positive control consists of one dilution of SVHR stock virus of known titer sufficient to give ~20 plaques. This number of plaques is significant statistically.
It is important that when going from a high concentration to a low concentration the pipet tip is changed to avoid “carry over” contamination. However, when pipetting from a low concentration to a higher concentration, as is done when the wells are inoculated, a single pipet tip can be used.
Materials and Reagents
Note: These can be from any supplier that offers Cell Culture grade materials or reagents except where specified. All culture flasks and plates are standard and can be from any supplier except where noted.
All fetal bovine serum (FBS) should be heat inactivated at 56 °C for 30 min.
- Sub-culture of vertebrate and invertebrate cells
Sub-culture of BHK-21 cells- 6-well plates
- Sterile tissue culture supplies (pipets, vented or plug cap flasks and 15 ml conical tubes)
- Heat-inactivated fetal bovine serum (FBS)
- Tryptose phosphate broth
- L-glutamine
- Gentamicin sulfate
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- MgCl2·6H2O
- Complete E-MEM, warmed to 37 °C (Earl’s salts minimal essential medium, completed with 10% heat inactivated fetal bovine serum, FBS)
- 1x PBS-D (see Recipes)
- Trypsin stock, 0.25%, warmed to 37 °C (see Recipes)
Sub-culture of C7-10 cells- Sterile tissue culture supplies (pipets, vented or plug cap flasks and 15 ml conical tubes)
- 70% ethanol
- 1x MEM
- Virus growth and titration
Stock virus preparation- Serological pipettes (5 ml, 10 ml and 25 ml)
- FBS (fetal bovine serum)
- Sterile Tissue culture grade water: for a detailed discussion of purified water and specifications for cell culture and other grades of water see Laboratory Water (The national institutes of health, March, 2013)
- Gentamicin sulfate
- L-glutamine
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- MgCl2·6H2O
- Neutral red
- Complete E-MEM, 1x and 2x (see Recipes)
- Gentamicin sulfate, 100x (mammalian cells only) (see Recipes)
- L-glutamine 100x (see Recipes)
- HEPES (pH 7.2-7.4), 1 M (see Recipes)
- 2% Neutral red stock solution (see Recipes)
- PBS-D (10x) (see Recipes)
- SVHR diluent (see Recipes)
- Tryptose phosphate broth (TPB) (optional for ATCC cells) (see Recipes)
Virus titration by plaque formation on BHK cells
BHK cells preparation- Sterile tissue culture supplies (pipets, vented or plug cap flasks and 15 ml conical tubes)
- Heat-inactivated fetal bovine serum (FBS)
- Tryptose phosphate broth
- L-glutamine
- Gentamicin sulfate
- Trypsin
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2
- CaCl2·2H2O
- KH2PO4
- MgCl2·6H2O
- Complete E-MEM, warmed to 37 °C (see Recipes)
- 1x PBS-D (see Recipes)
- Trypsin stock, 0.25%, warmed to 37 °C (see Recipes)
Virus titration by Plaque assay- 2% Agarose
- Heat inactivated Fetal Bovine Serum (FBS)
- 100% glycerol
- Heat-inactivated fetal bovine serum (FBS)
- Tryptose phosphate broth
- L-glutamine
- Gentamicin sulfate
- Trypsin
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- KH2PO4
- Neutral Red
- Complete E-MEM 2x (see Recipes)
- 2% Neutral Red (see Recipes)
- SVHR Diluent (see Recipes)
- 1x PBS-D (see Recipes)
- 1 M HEPES (pH 7.4) (see Recipes)
- Sindbis virus infection of vertebrate and invertebrate cells
Sindbis virus infection of BHK cells- Centrifuge tubes (Corning, 15 ml Fisherbrand, catalog number: 05-539-5)
- Sindbis virus infection of C7-10 cells
- 100% glycerol (sterile)
- Tissue culture grade water
- Heat-inactivated fetal bovine serum (FBS)
- Tryptose phosphate broth
- L-glutamine
- Gentamicin sulfate
- Trypsin
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- KH2PO4
- MgCl2·6H2O
- Tryptose phosphate broth (TPB; Difco)
- Complete E-MEM (see Recipes)
- Gentamicin sulfate 100x (see Recipes)
- L-glutamine 100x (see Recipes)
- 1 M HEPES (see Recipes)
- TPB-tryptose phosphate broth (see Recipes)
- PBS-D (10x) (see Recipes)
Sindbis virus infection of C7-10 cells- Glycerol
- Liquid N2
- Heat-inactivated fetal bovine serum (FBS)
- Tryptose phosphate broth
- L-glutamine
- Gentamicin sulfate
- Trypsin
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- KH2PO4
- MgCl2·6H2O
- Completed E-MEM medium (see Recipes)
- 1x PBS-D (see Recipes)
- Purification of Sindbis virus
Purification and Concentration by isopycnic centrifugation- Centrifuge tubes (ultra-clear)
- Small tube clamp
- Waste beaker
- 2 ml cryotube
- 15% potassium tartrate (dibasic, hemihydrate) in PBS-D (filter sterilize to store and store at 4 °C)
- 35% potassium tartrate in PBS-D (filter sterilize to store and store at 4 °C)
- BCA assay (PierceTM Rapid Gold BCA Protein Assay Kit) (Thermo Fisher Scientific, catalog number: A53225)
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- KH2PO4
- MgCl2·6H2O
- PBS-D pH 7.4 (see Recipes)
Concentration of virus by Polyethylene glycol (PEG) precipitation- Kimax high strength glass centrifuge tube (Kimax, catalog number: #Z2514888) (plain 30 ml)
- Polyallomer centrifuge tubes
- Polyethylene glycol-8000 (PEG-8000)
- 15% potassium tartrate (dibasic, hemihydrate) in PBS-D
- 35% potassium tartrate in PBS-D
- BCA assay (PierceTM Rapid Gold BCA Protein Assay Kit) (Thermo Fisher Scientific, catalog number: A53225)
- KCl
- KH2PO4
- NaCl
- Na2HPO4 or Na2HPO4·7H2O
- CaCl2 or CaCl2·2H2O
- KH2PO4
- MgCl2·6H2O
- Tris Buffer, pH 7.0
- EDTA
- PBS-D, pH 7.4 (see Recipes)
- PEG Buffer (see Recipes)
- 2 M NaCl (see Recipes)
- Calculation of particle to PFU ratio
None - Mass spectrometry and proteomic analysis
Note: All reagents should be LCV-MS grade.
Protein Extraction and Digestion- Lo-bind Centrifuge tubes (Eppendorf)
- Spin filter (Millipore-Ultracell YM-30)
- Pierce C18 spin columns (Thermo Fisher Scientific)
- Parafilm
- Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, catalog number: A53225)
- Mammalian Protein Extraction Reagent (M-PER) supplemented with 50 mM dithiothreitol (Thermo Fisher Scientific, catalog number: 78501)
- 0.05 M iodoacetamide in UA buffer
- Trypsin/Lys-C prepared in 100 mM TEAB to 10 µg/ml
- DTT
- CaCl2
- CaCl2·2H2O
- KH2PO4
- MgCl2·6H2O
- NaCl
- Urea
- Tris-HCl pH 8.5
- TEAB (Sigma, catalog number: T7408-100ML)
- 100% trifluoroacetic acid
- IAA powder
- Sterile 1x PBS (see Recipes)
- UA buffer (see Recipes)
- 100 mM triethylammonium bicarbonate (TEAB) (see Recipes)
- 0.5 M NaCl (see Recipes)
- 10% trifluoroacetic acid (TFA) (see Recipes)
- M-PER supplemented with 50 mM dithiothreitol (DTT)
- Iodoacetamide (IAA) solution (0.05 M IAA in UA buffer) (see Recipes)
- Trypsin digestion solution (see Recipes)
LC-MS/MS Data Acquisition- Picofrit 15 cm x 75 µm ID HPLC column packed with 5 µm BioBasic C18 particles 300Å (New Objective)
- 3% acetonitrile/0.1% formic acid
- 100% formic acid (Thermo Scientific, catalog number: 28905)
- 100% acetonitrile
- A Buffer (see Recipes)
- B Buffer (see Recipes)
Equipment
- Sub-culture of vertebrate and invertebrate cells
- 25 cm2 flask
- 75 cm2 flask
- CO2 Cell culture incubator (Any supplier is adequate)
Note: Any supplier is adequate; the choices are price/quality and capacity. All that is necessary is a water-jacketed incubator which can be regulated to 5% CO2 in a water-saturated atmosphere. This will require a medical grade CO2 tank (liquid gas under pressure) and a pressure regulator specific for that tank. Two incubators will be required if the cells are to be grown simultaneously because the vertebrate cells are incubated at 38 °C and invertebrate cells at 28 °C. - Biological safety cell culture hood
- Inverted microscope
- Hemacytometer
- Virus growth and titration
Stock virus preparation- CO2 Cell culture incubator
Virus titration by Plaque assay- Rocker platform (Bellco Biotechnology)
- 37 °C water bath
- Dilution tube rack
- Ice bucket
- Sindbis virus infection of vertebrate and invertebrate cells
Sindbis virus infection of BHK cells- Platform rocker (Bellco Biotechnology)
- Hemocytometer
- 75 cm2 flasks
- Centrifuge
- Hemocytometer
- Purification of Sindbis virus
Purification and Concentration by isopycnic centrifugation- Inverted microscope
- Platform rocker (Bellco Biotechnology)
- Hemocytometer or cell counting device
- CorningTM polypropylene tubes (micro centrifuge tubes, self-standing and conical)
- Sorvall RC-5B Super speed centrifuge
- Beckman ultracentrifuge
- Beckman SW-28 rotor
- Polyallomer 38 ml tubes
- Beckman SW-55Ti
- Polyallomer 5 ml tubes
- Ring stand
- Small tube clamp
- Hand held low-intensity lamp
- 30-100 ml gradient former (Bio-Rad, model 385)
- Medium stir plate
- Sorvall RC-5B Super speed centrifuge
- Beckman ultracentrifuge and appropriate rotor and tubes for the volume used
- 30-100 ml gradient former (Bio-Rad, model 385)
- Calculation of particle to PFU ratio
None - Mass spectrometry and proteomic analysis
Protein Extraction and Digestion- Tabletop centrifuge capable of reaching up to 14,000 x g
- Shaker heat block
- Vortex
- SpeedVac Concentrator
LC-MS/MS Data Acquisition- Orbitrap ELITE mass spectrometer (or equivalent; e.g., Orbitrap QE+, Orbitrap Fusion, Orbitrap Fusion Lumos)
- Easy-nLC II liquid chromatography system (or equivalent HPLC/UHPLC nanoflow pump system)
Software
Data Processing:
- High performance computer meeting the minimum specifications to run Proteome Discoverer Software (Currently available version: 2.2; Thermo Fisher Scientific)
- PANTHER classification system (http://www.pantherdb.org/)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Hernandez, R., Glaros, T., Rizzo, G. and Ferreira, D. F. (2019). Purification and Proteomic Analysis of Alphavirus Particles from Sindbis Virus Grown in Mammalian and Insect Cells. Bio-protocol 9(10): e3239. DOI: 10.21769/BioProtoc.3239.
分类
微生物学 > 微生物-宿主相互作用 > 病毒
微生物学 > 微生物蛋白质组学 > 全生物体
分子生物学 > 蛋白质 > 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












