- EN - English
- CN - 中文
Imaging Higher-order Chromatin Structures in Single Cells Using Stochastic Optical Reconstruction Microscopy
随机光学重建显微镜用于单细胞中高级染色体结构的成像
发布: 2019年02月05日第9卷第3期 DOI: 10.21769/BioProtoc.3160 浏览次数: 7595
评审: Zinan ZhouMichael Nguyen TrinhVishal S Parekh
Abstract
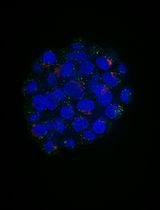
Higher-order chromatin organization shaped by epigenetic modifications influence the chromatin environment and subsequently regulate gene expression. Direct visualization of the higher-order chromatin structure at their epigenomic states is of great importance for understanding chromatin compaction and its subsequent effect on gene expression and various cellular processes. With the recent advances in super-resolution microscopy, the higher-order chromatin structure can now be directly visualized in situ down to the scale of ~30 nm. This protocol provides detailed description of super-resolution imaging of higher-order chromatin structure using stochastic optical reconstruction microscopy (STORM). We discussed fluorescence staining methods of DNA and histone proteins and crucial technical factors to obtain high-quality super-resolution images.
Keywords: Higher-order chromatin (高级染色体)Background
Recent advances in super-resolution imaging technique provide new potentials to observe the biological structures at the molecular scale. In particular, single-molecule localization based super-resolution technique such as stochastic optical reconstruction microscopy (STORM) has become a valuable tool to directly observe higher-order chromatin structure in situ down to ~20-30 nm resolution (Xu et al., 2018). Although many detailed protocols have been devoted for STORM imaging in general, few have been focused on imaging higher-order chromatin structures. Chromatin has densely packed structure, and the nucleus of mammalian cells tends to be thicker than other membrane-based proteins. Such samples tend to give higher background and present more overlapping fluorescent emitters. Therefore, the proper optimization of nuclear staining for STORM imaging is critical to obtain high-quality super-resolution images of chromatin structure. A compromise in any of staining steps may lead to significant image artifacts and degradation in image resolution. Here we provide detailed protocols for STORM-based super-resolution imaging of higher-order chromatin structure marked by either DNA and histone proteins. We also provide detailed procedures on the conjugation of the fluorophores with secondary antibodies, for single-color and two-color STORM imaging. The protocol presented here is optimized for STORM imaging, we believe that this protocol can also be extended to most high-resolution fluorescence imaging of chromatin structures or other proteins of interest.
Materials and Reagents
- Microcentrifuge tube
- Glass-bottom dish (World Precision Instruments, catalog number: FD3510)
- NAP-5 size-exclusion columns (GE Healthcare, catalog number: 17-0853-02)
- MCF-10A cells
- DMEM/F12 (Invitrogen, catalog number: 11039-021)
- Horse serum (Invitrogen, catalog number: 16050-122)
- Pen/Strep (100x solution) (Invitrogen, catalog number: 15070-063)
- EGF (Peprotech, 1 mg) (Resuspend at 100 μg/ml in sterile ddH2O. Store aliquots at -20 °C)
- Hydrocortisone (Sigma-Aldrich, catalog number: H0888) (Resuspend at 1 mg/ml in 200-proof ethanol and store aliquots at -20 °C)
- Cholera toxin (Sigma-Aldrich, catalog number: C-8052) (Resuspend at 1 mg/ml in sterile ddH2O and allow to reconstitute for about 10 min. Store aliquots at 4 °C)
- Insulin (Sigma-Aldrich, catalog number: I-1882) (Resuspend at 10 mg/ml in sterile ddH2O containing 1% glacial acetic acid. Shake solution and allow 10-15 min to reconstitute. Store aliquots at -20 °C)
- FluoSpheresTM Carboxylate-Modified Microspheres, 0.1 μm, yellow-green fluorescent (505/515) (Thermo Fisher Scientific, catalog number: F8803)
- TetraSpeckTM Microspheres, 0.1 μm, fluorescent blue/green/orange/dark red (Thermo Fisher Scientific, catalog number: T7279)
- Phosphate buffered saline (PBS) (Lonza)
- Triton X-100 (Sigma-Aldrich, catalog number: T8787)
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9647)
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148)
- Primary antibody
Rabbit anti H3K4me3 antibody (EMD Millipore, catalog number: 07-473)
Mouse anti H3K9ac antibody (Abcam, catalog number: ab12179) - Secondary antibodies
Donkey anti-rabbit antibody (Jackson ImmunoResearch, catalog number: 711-005-152)
Donkey anti-mouse antibody (Jackson ImmunoResearch, catalog number: 715-005-151) - Alexa 405 carboxylic acid succinimidyl ester (Thermo Fisher Scientific, catalog number: A30000)
- Alexa 647 carboxylic acid succinimidyl ester (Thermo Fisher Scientific, catalog number: A20006)
- Cy2 and Cy3B reactive dye (GE Healthcare, catalog numbers: PA22000, PA63101)
- Dimethylsulfoxide (DMSO; anhydrous)
- Click-iTTM EdU Alexa FluorTM 647 Imaging Kit (Thermo Fisher Scientific, catalog number: C10340)
- Glucose (Sigma-Aldrich, catalog number: G7021)
- Glucose oxidase from Aspergillus niger-Type VII (Sigma-Aldrich, catalog number: G2133)
- Catalase from bovine liver-lyophilized powder (Sigma-Aldrich, catalog number: C40)
- 1 M Tris, pH 8.0
- NaCl (Sigma-Aldrich, catalog number: S9888)
- 2-mercaptoethanol (β-ME) (Sigma-Aldrich, catalog number: 63698)
- Cysteamine (MEA) (Sigma-Aldrich, catalog number: 30070)
- NaHCO3 (Sigma-Aldrich, catalog number: S5761)
- Culture medium for MCF-10A (see Recipes)
- Washing buffer (see Recipes)
- Blocking buffer (see Recipes)
- STORM Imaging buffer (see Recipes)
Equipment
- Olympus IX71 inverted microscope frame with an oil-immersion objective (100x, NA = 1.4, UPLSAPO 100XO; Olympus, model: IX71)
- sCMOS camera (pco.edge 4.2, PCO-TECH)
Two-color dSTORM images were acquired on a custom system built upon an Olympus IX71 inverted microscope frame with an oil-immersion objective and an sCMOS camera attached on the side camera port. The 0.5x adaptor was used, such that each pixel on the camera corresponds to 130 nm on the sample plane (the pixel size of approximately 80-160 nm is acceptable). - Objective nanopositioner (Mad City Labs, model: Nano-F100S)
- N-STORM system (Nikon Instruments)
- NanoDrop 2000 microspectrophotometer (Thermo Fisher Scientific, model: NanoDropTM 2000)
- Rocking platform
Software
- ImageJ (National Institutes of Health, https://imagej.nih.gov/ij/download.html)
- ThunderSTORM plug in Ovesný et al. (2014) (downloadable at https://github.com/zitmen/thunderstorm)
- Labview (National Instrument)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Xu, J. and Liu, Y. (2019). Imaging Higher-order Chromatin Structures in Single Cells Using Stochastic Optical Reconstruction Microscopy. Bio-protocol 9(3): e3160. DOI: 10.21769/BioProtoc.3160.
分类
细胞生物学 > 细胞成像 > 荧光
分子生物学 > DNA > DNA 结构
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link