- EN - English
- CN - 中文
Assessing Yeast Cell Survival Following Hydrogen Peroxide Exposure
过氧化氢胁迫条件下酵母细胞存活率的评估
发布: 2019年01月20日第9卷第2期 DOI: 10.21769/BioProtoc.3149 浏览次数: 11867
评审: Emily CopeLionel SchiavolinAnonymous reviewer(s)
Abstract
In the presence of oxidative stress, cellular defense systems that can detoxify reactive oxygen species are activated through multiple signaling cascades and transcriptional reprogramming. The budding yeast Saccharomyces cerevisiae has served as an excellent model for genetically-identifying factors important for the response to oxidative stress. Here, we describe two assays for testing yeast gene deletion strains or strains overexpressing a gene of interest for viability following oxidative stress induced by hydrogen peroxide treatment. These include a plate-based spot assay for visualizing cell growth and a quantitative colony counting assay. As stress response assays can be highly variable depending on cell growth conditions, these protocols have been optimized for obtaining highly-reproducible results between experiments. We demonstrate the use of these protocols for genetic tests of a putative chromatin regulator implicated in regulating the transcriptional response to oxidative stress.
Keywords: Yeast (酵母)Background
The unicellular eukaryote Saccharomyces cerevisiae has been an exceptional model for revealing large networks of genes critical to the survival of cells in diverse types of environmental stress (Gasch et al., 2000; Weiner et al., 2012; Ho and Gasch, 2015), largely due to the excellent genetic tools available in this system and the ease with which stress responses can be evaluated. In particular, genetic studies in yeast have uncovered the regulatory mechanisms that control the cells’ response to oxidative stress (Morano et al., 2012). Environmental factors, including radiation and the presence of redox-cycling agents, are a source of oxidative stress which cause increased reactive oxygen species (ROS). These ROS include the superoxide anion, H2O2, and hydroxyl radicals, which are highly reactive with proteins, lipids, and nucleic acids, causing oxidative damage to all of these macromolecules (Schieber and Chandel, 2014). Aerobic respiration also generates low levels of endogenous ROS by the leakage of electrons from the electron transport chain, and mitochondrial dysfunction, as occurs with stress or aging, increases ROS levels further, which triggers the induction of cellular defense mechanisms such as activated expression of antioxidant enzymes which detoxify ROS (Schieber and Chandel, 2014). Pathologies including cancer, cardiovascular disease, autoimmune diseases, and aging are associated with deregulated oxidative stress defense systems and altered levels of intracellular ROS. Therefore, identifying and characterizing the network of proteins responsible for protecting cells during oxidative stress is key to defining the pathophysiology underlying these diseases.
To test the role of a given gene in the response to oxidative stress, it is advantageous to analyze yeast strains which are deleted or otherwise mutated for the gene and which overexpress the protein encoded by the gene. If a particular gene is required to protect cells during oxidative stress, it is expected that its deletion would render cells sensitive to the stress, and its overexpression would cause resistance to the stress. The assays described below test the survival of yeast cells following treatment with hydrogen peroxide, which has served as a model compound for inducing oxidative stress in yeast (Morano et al., 2012; Kwolek-Mirek and Zadrag-Tecza, 2014). Specifically, these protocols were used to test the role of the chromatin regulator Set4 during oxidative stress (Tran et al., 2018). We used a set4∆ yeast strain, and a strain carrying a plasmid for the inducible overexpression of SET4 in the presence of β-estradiol, as previously described (McIsaac et al., 2013). The raw data shown here were obtained to generate the results published in Tran et al. (2018) that demonstrate a role for Set4 in protecting cells during oxidative stress.
We present both a semi-quantitative plate spot assay and a quantitative colony forming unit (cfu) survival assay to interrogate the survival of cells following hydrogen peroxide treatment. We found it useful to be able to visualize the growth differences in cells on the plates and validate these differences quantitatively using the cfu assay. Both protocols here appeared to show much more consistent results than plate spot assays in which hydrogen peroxide is added to the medium in the plate prior to spotting the yeast on the plate (Kwolek-Mirek and Zadrag-Tecza, 2014). In addition, we applied methods to optimize the growth conditions and hydrogen peroxide concentration to account for the observations that yeast cells acquire resistance to hydrogen peroxide when grown in nutrient-poor media, such as synthetic complete or dropout media, or following the diauxic shift, which occurs when cells transition from primarily glucose metabolism via glycolysis to aerobic respiration of ethanol (Guan et al., 2012). When these conditions are not accounted for, results are highly variable and may represent confounding factors in the experiment, rather than the consequences of the deletion or overexpression of the gene being investigated. These assays are not resource-intensive and can be used to test multiple mutant or overexpressing strains simultaneously. Overall, these protocols represent a sensitive and highly-reproducible approach to assaying the response of yeast cells to oxidative stress.
Materials and Reagents
- Pipette tips
- P2 tips (Gilson, catalog number: F171200)
- P200 tips (Gilson, catalog number: F171300)
- P1000 tips (Gilson, catalog number: F171500)
- Toothpicks and inoculating sticks (sterilized) (VWR, catalog number: 12000-806)
- Petri dish (Corning, catalog number: 351029)
- Assay Plate, 96-well, U bottom (Falcon, catalog number: 62406-015)
- 1.5 ml microfuge tubes (Fisherbrand, catalog number: 05-408-129)
- yEG001
- yEG322
- yEG315
- yEG372
- yEG375
- 50 ml conical centrifuge tubes (Falcon, catalog number: 14-432-22)
- Yeast strains (Table 1)
- Vector pMN3 (empty vector) (McIsaac et al., 2013)
- Vector pMN3-SET4 (Tran et al., 2018)
- Sterile, distilled water
- Yeast extract (Research Products International, catalog number: Y20025)
- Peptone (Research Products International, catalog number: P20250)
- Dextrose (Thermo Scientific, catalog number: BP350-1)
- Yeast nitrogen base (US Biologicals, catalog number: Y2025)
- Drop-out Mix Synthetic Minus Uracil (US Biologicals, catalog number: D9535)
- Ethanol (Sigma-Aldrich, catalog number: E7023)
- Hydrogen peroxide (Fisher Scientific, catalog number: H325-100)
- β-estradiol (Millipore Sigma, catalog number: E2758-1G)
- Agar (US biological, catalog number: A0930)
- YPD (Yeast extract-Peptone-Dextrose) medium (see Recipes)
- Synthetic Complete without Uracil (SC-URA) medium (see Recipes)
- 10 mM β-estradiol (see Recipes)
Table 1. Yeast strains used in this protocol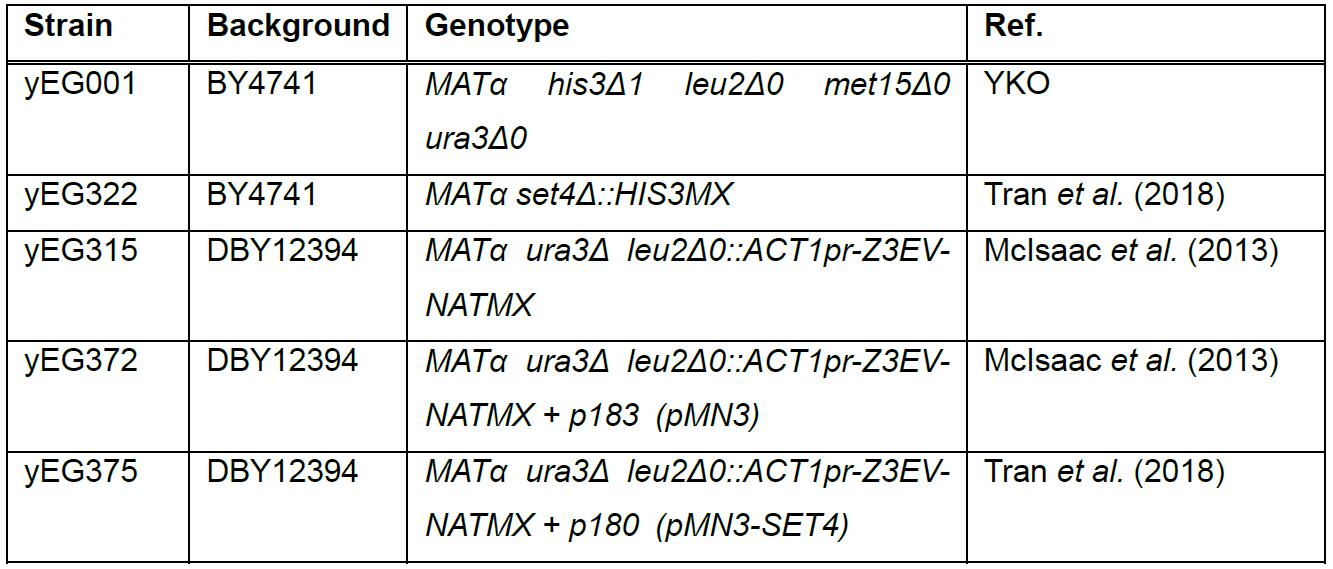
Equipment
- 125 ml (or 50 ml, if available) Erlenmeyer flasks (VWR, catalog number: 10536-912)
- Glass cell spreaders (VWR, catalog number: 89166-788)
- Single-channel pipettors
- P2 pipette (Gilson, catalog number: F144054M)
- P20 pipette (Gilson, catalog number: F144056M)
- P200 pipette (Gilson, catalog number: F144058M)
- P1000 pipette (Gilson, catalog number: F144059M)
- Glass culture tubes (15 ml) (Corning, catalog number: 982016X)
- Multichannel Pipettors (Pipetman Neo P8 x 20N and Pipetman Neo P8 x 200N)
- Incubator
- Rollordrum Tc-7 Tissue Culture Rotator (New Brunswick Scientific)
- Incubator Shaker (New Brunswick Scientific G-25, temperature required: 30 °C; shaker dimensions: 22 in. x 45.5 in. x 30.5 in)
- Vortexer (VWR Standard Heavy Duty Vortex Mixer, catalog number: 97043-562)
- Spectrophotometer (Thermo Scientific, model: NanoDrop 2000c, catalog number: ND-2000c)
- MultiDoc-it Imaging system (UVP)
- Centrifuge (Beckman Coulter Allegra X-14R, catalog number: A99465)
- Microfuge (Beckman Coulter Microfuge 20R)
- Autoclave (Steris AMSCO Century SG-120 Scientific Gravity Sterilizer)
Software
- Excel, Microsoft Office
- Prism GraphPad
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Tran, K. and Green, E. M. (2019). Assessing Yeast Cell Survival Following Hydrogen Peroxide Exposure. Bio-protocol 9(2): e3149. DOI: 10.21769/BioProtoc.3149.
- Tran, K., Jethmalani, Y., Jaiswal, D. and Green, E. M. (2018). SET4 is a chromatin-associated protein, promotes survival during oxidative stress, and regulates stress response genes in yeast. J Biol Chem 293(37): 14429-14443.
分类
微生物学 > 微生物细胞生物学 > 细胞活力
微生物学 > 微生物遗传学 > 基因表达
细胞生物学 > 细胞活力 > 细胞存活
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













