- EN - English
- CN - 中文
Visualization and Quantification of Cell-to-cell Movement of Proteins in Nicotiana benthamiana
本氏烟中蛋白的细胞间运动的可视化定量检测
发布: 2018年12月20日第8卷第24期 DOI: 10.21769/BioProtoc.3114 浏览次数: 7634
评审: Zhibing LaiChao-Jan LiaoJuan Du
Abstract
Cell-to-cell movement of proteins through plasmodesmata is a widely-established mechanism for intercellular signaling in plants. Current techniques to study intercellular protein translocation rely on single-cell transformation using particle bombardment or transgenic lines expressing photo-inducible fluorophores. The method presented here allows visualization and objective quantification of (effector) protein movement between N. benthamiana leaf cells. Agroinfiltration is performed using a single binary vector encoding a GFP-tagged protein of interest that is either mobile or non-mobile (MP; non-MP), together with an ER-anchored mCherry. Upon creation of mosaic-like transformation patterns, cell-to-cell movement of the MP can be followed by monitoring translocation of the GFP signal from mCherry labeled transformed cells into neighboring non-transformed cells. This process can be visualized using confocal microscopy and quantified following protoplast isolation and flow cytometric cell analysis. This method overcomes the limitations of existing methods as it allows rapid and objective quantification of protein translocation without the need of creating transgenic plants.
Keywords: Protein translocation (蛋白迁移)Background
Many proteins use plasmodesmata as channels to move from one cell to another. The aperture of plasmodesmata determines the size of molecules able to pass. To visualize whether a protein is able to move from cell to cell, current techniques include particle bombardment and photo-inducible fluorophores (e.g., DRONPA) (Gerlitz et al., 2018). A drawback of DRONPA is that it is highly time consuming since transgenic lines have to be created (Gerlitz et al., 2018). Transient transformation using particle bombardment overcomes this limitation. However, since it involves single cell transformation the fluorescent signal fades out rapidly when distributed over neighboring cells (Figure 1A). Fading is a limitation of both techniques and hampers detection of movement of poorly expressed proteins (Cao et al., 2018). Moreover, quantification of protein translocation with these methods is laborious and subjective, as only relative low numbers of translocation events can be monitored using a confocal microscope.
Our newly developed method allows both visualization and objective quantification of intercellular protein translocation in N. benthamiana leaves. The method is derived from a protocol for analyzing cell-to-cell movement of the Non-Structural Movement (NSm) protein from Tomato Spotted Wilt Virus (Feng et al., 2016). By transiently transforming multiple cells in a mosaic-like pattern movement of green fluorescently tagged proteins from transformed (labeled red) to non-transformed cells (non-fluorescent) can be monitored (Figure 1B). We adjusted this method to allow monitoring of effector translocation and provide an unbiased quantification procedure to record cell-to-cell movement. The effector proteins Avr2 and Six5 from the fungus Fusarium oxysporum were employed to test this system. This effector pair manipulates plasmodesmata to facilitate cell-to-cell movement of Avr2 in a Six5 dependent manner (Cao et al., 2018). In the absence of Six5 the Avr2 protein behaves as a non-mobile protein (Non-MP) and is retained in the transformed cell. The dependency of one protein on another one for cell-to-cell movement represents an anomaly, as typically mobile proteins (MP) have intrinsic properties to translocate themselves as exemplified by the NSm protein (Feng et al., 2016). 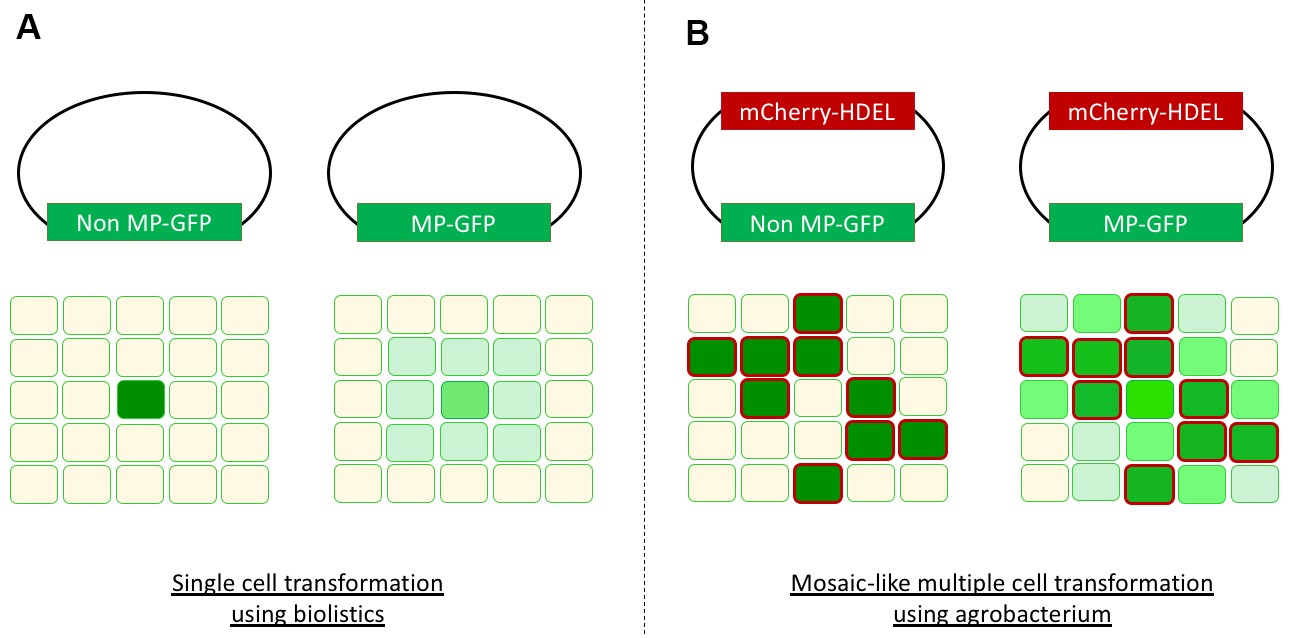
Figure 1. Overview cell-to-cell movement visualization. A. Single cell transformation using biolistics with a non-mobile protein (Non-MP-GFP) will result in a single green cell (left). A mobile protein (MP-GFP) will spread to neighboring cells, resulting in a green halo (right). B. Agrobacterium-mediated transient transformation of multiple cells in a mosaic-like pattern. In case of a non-mobile protein the transformed cells are marked both red (mCherry-HDEL) and green (Non-MP-GFP), and surround non-fluorescent untransformed cells (left). A mobile protein (MP-GFP) will be translocated from transformed cells (mCherry-HDEL and MP-GFP) to the neighboring untransformed cells resulting in an increased GFP signal in these cells (right). The intensity of this GFP signal in the untransformed cells varies depending on the amount of neighboring transformed cells, an artist’s impression is depicted here.
For our study a single binary vector carrying both Avr2-GFP and mCherry-HDEL was used for agroinfiltration in N. benthamiana (Ma et al., 2012). The mCherry-HDEL protein is ER-localized and cannot translocate to neighboring cells, thereby serving as a marker for transformed cells. Transformed cells show both green GFP and red mCherry signals, while untransformed cells lack both signals. However, upon co-infiltration with a second binary vector encoding SIX5 these neighboring untransformed cells will become green fluorescent as they acquire the AVR2-GFP protein from neighboring transformed cells. To quantify the intercellular translocation of Avr2-GFP, protoplasts can be isolated from infiltrated leaf sectors and subjected to flow cytometric cell analysis (Figure 2). Flow cytometry allows determination of the number of GFP + mCherry and GFP-only cells in presence- and absence of Six5, thereby enabling quantification of AVR2-GFP effector translocation. Potential false positive GFP-only events will be recorded in the AVR2-GFP control lacking SIX5, allowing determination of the base line level of GFP-only cells. Use of flow cytometry to quantify cell-to-cell movement provides an objective and unbiased way to assess intercellular translocation in a large number of cells. Other possible applications for this method include, among others, plasmodesmata aperture measurement by using single, double or triple GFP fusions following treatment of the plant with various (a)biotic stresses and/or pharmaceutical compounds. Furthermore, mutants and derivatives of proteins-of-interest can be screened for their altered translocation properties. 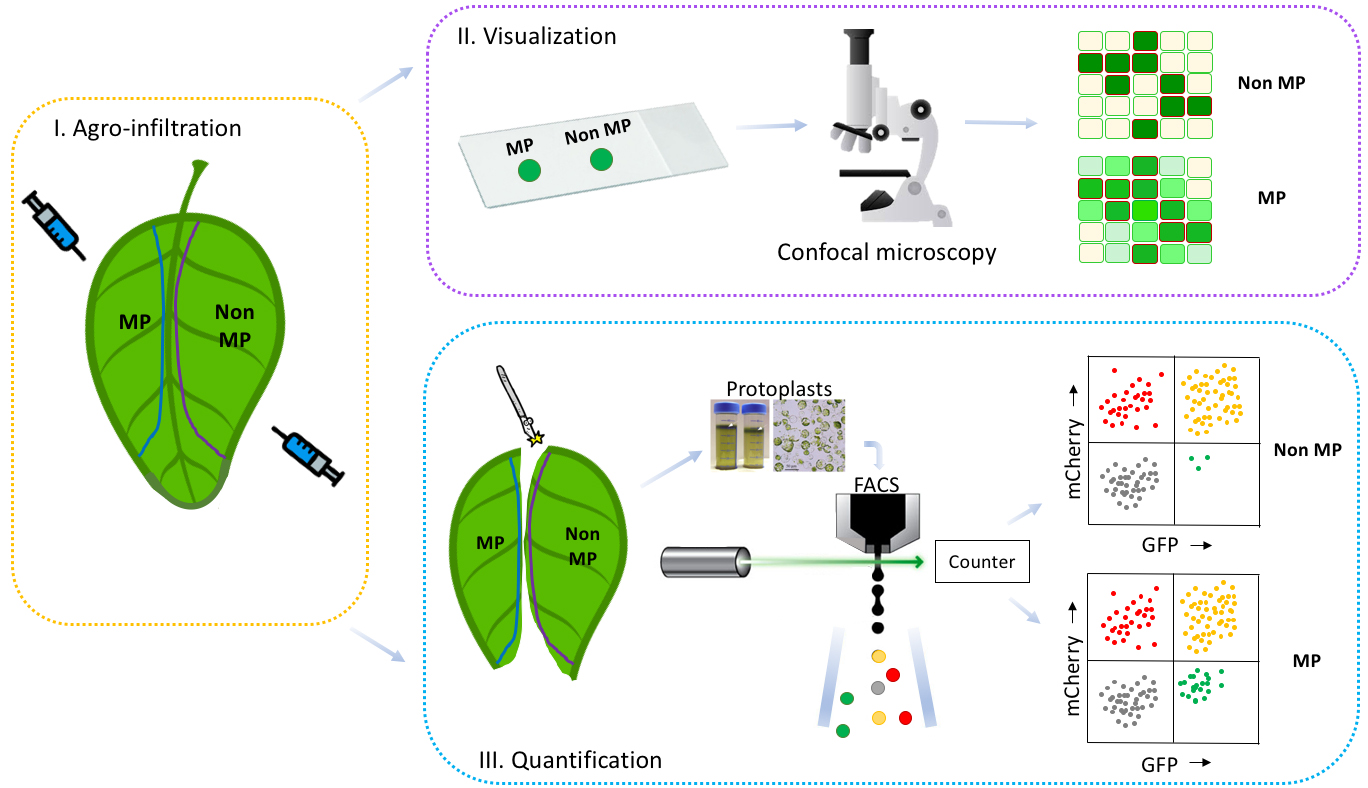
Figure 2. Workflow of the method to detect and quantify cell-to-cell movement of proteins. A schematic representation of the workflow (I, Agro-infiltration; II, Visualization; III, Quantification) to determine cell-to-cell movement of proteins is shown. By using this method to study the mobility of a protein of interest, one of two potential patterns will be obtained: either a mobile protein pattern (II, III MP) or a non-mobile protein pattern (II, III Non-MP). By comparing the protein of interest’s pattern with a positive MP control and a negative Non-MP control, a conclusion can be drawn regarding the protein of interest mobility. Our workflow consists of the following three steps: I. Infiltrate sectors of N. benthamiana leaves with Agrobacterium harboring constructs containing genes encoding either mobile (MP) or non-mobile proteins (Non-MP). II. Harvest leaf disks from the infiltrated leaf sectors and analyze the mosaic-like transformation patterns for MP and Non-MP with confocal microscopy to detect potential cell-to-cell movement. III. Isolate protoplasts from the infiltrated leaf sectors and count the number of red, yellow or green colored cells with flow cytometry (FACS) to quantify cell-to-cell movement.
Materials and Reagents
- Syringes 1 ml (BD Plastikpak, catalog number: 303172)
- Centrifuge tube 50 ml, conical bottom (Greiner Bio One, catalog number: 210261)
- Plastic pipette 25 ml (Greiner Bio One, catalog number: 760180)
- Microscope slides (Thermo Scientific, Menzel-Glazer, catalog number: 631-0098)
- Cover slides, 24 x 60 mm (Thermo Scientific, Menzel-Glazer, catalog number: 15747592)
- Square plates, 120 mm (Greiner Bio One, catalog number: 688102)
- Petri dishes, 94 mm (Greiner Bio One, catalog number: 632102)
- Non-sterile scalpel blades, No. 21 curved cutting edge (Swann-Morton, catalog number: 0107)
- Aluminum foil, thin
- Glass Pasteur pipettes (long) (WU Mainz, catalog number: 10216234)
- Steel mesh 80 (177 μm) (cell culture reagents) (Sigma, catalog number: S3770)
- 5 ml Polypropylene round bottom tubes (Falcon, catalog number: 352063)
- Membrane filter, 0.45 μm pore size (Millipore, Milex-HV, catalog number: SLHV033RS)
- Edding 400 permanent marker, 1 mm round point (Edding)
- E. coli (DH5α)
- Agrobacterium (GV3101)
- Nicotiana benthamiana
- pZK538 vector (Addgene, catalog number: 118012)
- pDGB3alpha2_35S:P19:Tnos (GB1203) (p19) (Addgene, catalog number: 68214)
- Avr2-GFP pZK538 (Addgene, catalog number: 118011) (Cao et al., 2018)
- NSm pZK538 (Addgene, catalog number: 118011-13) (Feng et al., 2016)
- Phusion polymerase
- DMSO
- Gamborg B5 medium (Duchefa, catalog number: G0209)
- MES sodium salt (Duchefa, catalog number: M1503)
- Calcium chloride dihydrate (CaCl2•2H2O) (Merck, catalog number: 102382)
- Ammonium nitrate (NH4NO3) (Merck, catalog number: 101187)
- Sucrose (Duchefa, catalog number: S0809)
- Macerozyme R-10 (Duchefa, catalog number: M8002)
- Cellulase onozuka RS (Duchefa, catalog number: C8003)
- Ethanol (96%) (VRW chemicals, catalog number: 83804)
- Commercial bleach (< 5% Hypochlorite)
- Sterile MilliQ water
- Potassium hydroxide (KOH) (Merck, catalog number: 105033)
- DpnI restriction enzyme (Thermo Scientific, catalog number: ER1701)
- GeneJet PCR purification kit (Thermo Scientific, catalog number: K0702)
- GeneJet Gel extraction kit (Thermo Scientific, catalog number: K0692)
- NEBuilder Hifi DNA assembly cloning kit (NEB, catalog number: E5520S)
- Agarose (Sphaero Q, catalog number: s103a)
- MS basal salt mixture (Duchefa, catalog number: M0221)
- Acetosyringone (C10H12O10) (Merck, catalog number: D134406)
- TEX buffer (see Recipes)
- Leaf digestion solution (see Recipes)
- MMAi medium (see Recipes)
Equipment
- Scalpel holder
- Scale
- Burner
- Flow hood
- Forceps
- Pipettes
- Leaf puncher 5 mm (Well Tech, Rapid core, catalog number: E69039-50)
- Confocal microscope LSM 510 (Zeiss, model: LSM510)
- EVOS FL cell imaging system (Thermo Fisher, model: EVOS FL, catalog number: AMF4300)
- Haemocytometer 0.100 mm; 0.0025 m2 (Merck, Blaubrand, catalog number: BR719520)
- Rotina 420R Centrifuge (Hettich Zentrifugen, model: Rotina 420R, catalog number: 4701)
- PCR machine (Bio metra, TAdvanced)
- Gel electrophoresis machine (Bio-Rad)
- Pipetus pipetting pump (Hirschmann Laborgerate, Pipetus, catalog number: 9907200)
- Heat block (Grant Instruments, model: QBD2, catalog number: 11470958)
- FACS Aria III BD (BD Biosciences, model: FACS Aria III)
Software
- Kaluza Flow Cytometry Analysis software version 1.2 (Beckman Coulter, US)
- Fiji (Fiji is just ImageJ) version 1 (https://fiji.sc/) (Schindelin et al., 2012)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Blekemolen, M. C., Tark-Dame, M. and Takken, F. L. (2018). Visualization and Quantification of Cell-to-cell Movement of Proteins in Nicotiana benthamiana. Bio-protocol 8(24): e3114. DOI: 10.21769/BioProtoc.3114.
分类
植物科学 > 植物细胞生物学 > 细胞间通讯
植物科学 > 植物转化 > 农杆菌介导的转化方法
分子生物学 > 蛋白质 > 细胞间转移
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link