- EN - English
- CN - 中文
Generation of Gene Knockout and Gene Replacement with Complete Removal of Full-length Endogenous Transcript Using CRISPR-Trap
通过CRISPR-Trap完全移除内源全长转录本进行基因敲除和基因置换
(*contributed equally to this work) 发布: 2018年10月20日第8卷第20期 DOI: 10.21769/BioProtoc.3052 浏览次数: 9390
评审: Renate WeizbauerIndranil MalikMolly Leung
Abstract
This protocol describes the application of the CRISPR-Trap from designing of the gene targeting strategy to validation of successfully edited clones that was validated on various human cell lines, among them human induced pluripotent stem cells (hiPSCs). The advantage of CRISPR-Trap over conventional approaches is the complete removal of any endogenous full-length transcript from the target gene. CRISPR-Trap is applicable for any target gene with no or little coding sequence in its first exon. Several human cell lines and different genes have so far been edited successfully with CRISPR-Trap.
Keywords: CRISPR (CRISPR)Background
The advent of CRISPR/Cas9 technology facilitated the genomic targeting for the generation of gene knockouts and gene editing. The conventional method to perform a knockout relies on the introduction of a frameshift leading to premature termination codons (PTCs), truncating the open reading frame (ORF) and subsequent degradation of the transcript of the targeted gene by nonsense-mediated mRNA decay (NMD). A possible pitfall of this approach is full-length transcripts which may escape NMD and give rise to C-terminal truncated proteins harboring residual or even dominant negative functions. This protocol presents the CRISPR-Trap, a method we recently established (Reber et al., 2018), which upon successful editing will prevent the expression of any full-length transcript from the target gene locus (Figure 1). Simply put, this approach targets the first intron of the gene of interest with CRISPR/Cas9. Using homology-directed repair (HDR) a customizable cassette flanked by a strong 3’-splice site and a strong polyadenylation signal is introduced in the first intron, thereby generating an artificial second and effectively last exon. Since transcription is terminated by the introduced polyadenylation signal, only the first endogenous exon and the inserted cassette is transcribed. The customizable cassette can be used to introduce a selection marker, thereby enabling easy selection for at least heterozygous edited clones. If a gene replacement is wanted, the customizable cassette can be used to introduce the replacement gene, followed by an internal ribosomal entry site (IRES) and a selection marker (Reber et al., 2016 and 2018).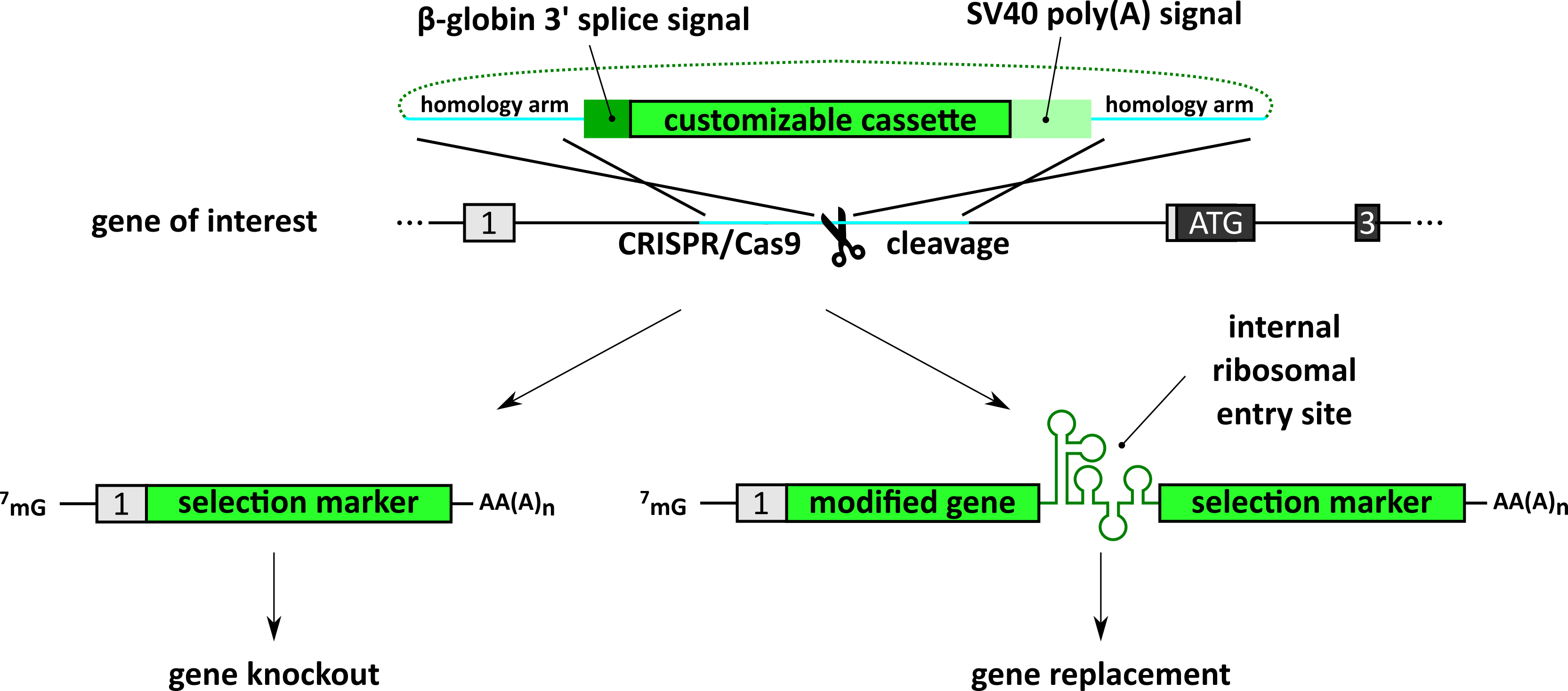
Figure 1. Schematic of the application of the CRISPR-Trap. The first intron of the target gene is cleaved using the CRISPR/Cas9 system and template DNA for homology-directed repair is provided. The template DNA contains a strong 3’ prime splice signal (dark green), a customizable cassette (light green) and a strong polyadenylation signal (turquoise). The customizable cassette can be utilized to either knockout (left) or replace the target gene (right). The cassette contains a selection marker that will be under the control of the endogenous promoter of the target gene upon successful editing. For gene replacements, an IRES is introduced in between the replacement gene and the selection marker. Figure adapted from Reber et al. (2018).
Materials and Reagents
- Pipette tips
- 10 µl sapphire bulk non-sterile pipette tips (Greiner Bio One International, catalog number: 771250 )
- 200 µl polypropylene universal pipette tips with graduation (Greiner Bio One International, catalog number: 739282 )
- 1,250 µl sapphire bulk non-sterile pipette tips (Greiner Bio One International, catalog number: 750250 )
- TPP® tissue culture plates
- TPP cell scraper 20 cm (MIDSCI, catalog number: TP 99010 )
- Cloning cylinder sterile, 3/16’’ ID x 5/16’’ H (SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: 37847-0100 )
- Cell strainer (EASYstrainer, 70 µm) (Greiner Bio One International, catalog number: 542070 )
- Bel-ArtTM SP SciencewareTM BelpenTM black markers (SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F13374-0000 )
- Plasmid of choice, serving as homology directed repair donor template (donor plasmid)
- Plasmid of choice for Cas9 and sgRNA expression, e.g., pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene, catalog number: 42230 )
- Transformation competent E. coli strain of choice for cloning and plasmid amplification (e.g., XL10 Gold) (Agilent, catalog number: 200314 )
- Transfection reagent of choice
- Cell detachment solutions
- hiPSCs
- Other human cell lines: Trypsin-EDTA (0.05%), phenol red (Thermo Fisher Scientific, catalog number: 25300054 )
- hiPSCs
- To grow hiPSCs as single cells: Y-27632, RHO/ROCK pathway inhibitor (STEMCELL Technologies, catalog number: 72302 )
- ZeocinTM (InvivoGen, catalog number: ant-zn )
- Puromycin dihydrochloride (Santa Cruz Biotechnology, catalog number: sc-108071A )
- PBS (Ca2+, Mg2+ free) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- TRIZOL (Sigma-Aldrich, catalog number: T9424 )
- Dow Corning® High-Vacuum silicone Grease (Sigma-Aldrich, catalog number: Z273554 )
- TOPOTM TA CloningTM Kit, Dual Promoter (Thermo Fisher Scientific, catalog number: 450640 )
- Chloroform for analysis EMSURE® ACS, ISO, Reag. Ph Eur (Merck, catalog number: 102445 )
- Absolute EtOH (Sigma-Aldrich, catalog number: 59176 )
- Trisodium citrate (Sigma-Aldrich, catalog number: W302600 )
- 2-propanol for analysis EMSURE® ACS, ISO, Reag. Ph Eur (Merck, catalog number: 109634 )
- RNase free glycogen (Thermo Fisher Scientific, catalog number: R0551 )
- DNA extraction kit (Quick-DNATM miniprep Kit) (ZYMO RESEARCH, catalog number: D3025 )
- NaOH (Sigma-Aldrich, catalog number: 306576 )
- Maximo Taq DNA Polymerase 2X-preMix/PCR Master Mix (GeneON, catalog number: S113 )
- KAPA Taq ReadyMix PCR Kit (KAPA Biosystems, catalog number: KK1006)
Equipment
- Pipettes
- Mammalian culture equipment
- Heat block (VWR, catalog number: 444-0938 )
- Thermocycler (VWR, catalog number: 732-2551 )
- NanoDrop (Thermo Fisher Scientific, model: NanoDropTM 2000 , catalog number: ND-2000)
- Cooling Centrifuge (Eppendorf, model: 5424 R , catalog number: 5404000010)
- Wide-field microscope
- Vortex (Scientific Industries, catalog number: SI-0266 )
Software
- Clone manager (http://www.scied.com/pr_cmpro.htm) or other cloning software
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Mechtersheimer, J., Reber, S. and Ruepp, M. (2018). Generation of Gene Knockout and Gene Replacement with Complete Removal of Full-length Endogenous Transcript Using CRISPR-Trap. Bio-protocol 8(20): e3052. DOI: 10.21769/BioProtoc.3052.
分类
分子生物学 > DNA > 诱/突变
分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link