- EN - English
- CN - 中文
Microtitre Plate Based Cell-SELEX Method
基于微量滴定板的Cell-SELEX技术
(*contributed equally to this work) 发布: 2018年10月20日第8卷第20期 DOI: 10.21769/BioProtoc.3051 浏览次数: 7950
评审: Longping Victor TseVaibhav B. ShahAnonymous reviewer(s)
Abstract
Aptamers have emerged as a novel category in the field of bioreceptors due to their wide applications ranging from biosensing to therapeutics. Several variations of their screening process, called SELEX have been reported which can yield sequences with desired properties needed for their final use. We report a facile microtiter plate-based Cell-SELEX method for a gram-negative bacteria E. coli. The optimized protocol allows the reduction of number of rounds for SELEX by offering higher surface area and longer retention times. In addition, this protocol can be modified for other prokaryotic and eukaryotic cells, and glycan moieties as target for generation of high affinity bio-receptors in a short course of time in-vitro.
Keywords: Aptamers (适配子)Background
Aptamers are single strands of synthetic DNA or RNA described in 1990 (Ellington and Szostak, 1990; Tuerk and Gold, 1990) with a distinct 3D geometry, which is a manifestation of their sequences. Different sequences allow the synthesis of aptamers, which can bind to an array of molecules ranging from small molecules to large proteins. This allowed aptamers to emerge as the rivals to conventional antibodies, which are limited to proteins as their targets due to structural limitations and can not be raised against high-risk pathogens as these usually kill the host much earlier than the time required to generate high affinity antibodies. Aptamers are generated in in-vitro setups and thus can effectively be used for high-risk pathogens. SELEX (Systematic Evolution of Ligands by EXponential enrichment) is an iterative process involving repeated exposures and separation leading to screening of a pool with dissociation constants in low nM ranges. Several variations of SELEX have been proposed which use a variety of stationary matrices including capillary electrophoresis SELEX, affinity chromatography based SELEX, magnetic bead SELEX, in-vivo SELEX, FACS SELEX and Microfluidics based SELEX, etc. These methods present various advantages over others but the number of SELEX rounds are consistently high.
A standard SELEX method has four major steps; exposure of random sequence single stranded nucleotide oligo library to target, binding of oligos to target molecule, selection of binders and removal of non-binding oligos, amplification of the binder fraction and portioning of the amplicon to single strand. These steps are iteratively performed till pool of high binding aptamer are screened out from the library. The oligonucleotide library consists of a random base-sequence flanked on both ends by primer binding sites, which aids in amplification and enrichment (Safeh et al., 2010; Kim et al., 2013).
We report a microtiter plate-based Cell-SELEX method, which can be employed over a wide number of molecules and cell types (Figure 1). The advantage proposed by the method is uniquely low number of rounds, which are the outcome of large surface area, higher retention times and a novel superior bio-probe based partitioning process (Priyanka et al., 2014).
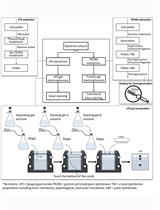
Figure 1. Concise Protocol Schematic. A step-by-step flowchart showing the various steps of Cell-SELEX. (Modified from author’s work Kaur et al., 2017)
Materials and Reagents
- Pipette tips, 10 μl (Tarsons, catalog number: 523100 )
- Pipette tips, 200 μl (Tarsons, catalog number: 523101 )
- Pipette tips, 1 ml (Tarsons, catalog number: 523104 )
- Nunc Maxisorp F96 microtitre plates (Thermo Fisher Scientific, catalog number: 437111 )
- Centrifuge tubes 50 ml (Tarsons, catalog number: 546041 )
- Microcentrifuge tubes (MCT) 1.5 ml (Tarsons, catalog number: 500010 )
- PCR Tubes 0.2 ml (Tarsons, catalog number: 510051 )
- Bacterial strains (E. coli)
- ssDNA library and primers (IDT custom synthesis):
Forward Primer: 5'-ATCCAGAGTGACGCAGCA-3'
Reverse Primer: 5'-biotin-ACTAAGCCACCGTGTCCA-3'
Library: 5'-ATCCAGAGTGACGCAGCA-45N-TGGACACGGTGGCTTAGT-3' - LB broth (HiMedia Laboratories, catalog number: M1245 )
- Phenylboronic acid (HiMedia Laboratories, catalog number: RM1599 )
- Streptavidin-Gold from Streptomyces avidinii (Sigma-Aldrich, catalog number: 53134 )
- Tris-HCl (HiMedia Laboratories, catalog number: MB030 )
- Glycine (HiMedia Laboratories, catalog number: MB013 )
- Magnesium chloride (HiMedia Laboratories, catalog number: MB040 )
- Sodium chloride (HiMedia Laboratories, catalog number: MB023 )
- Sodium phosphate monobasic (HiMedia Laboratories, catalog number: GRM3963 )
- Sodium phosphate dibasic (HiMedia Laboratories, catalog number: MB024 )
- Tris base (HiMedia Laboratories, catalog number: TC072 )
- Sodium carbonate (HiMedia Laboratories, catalog number: GRM851 )
- Sodium bicarbonate (HiMedia Laboratories, catalog number: TC230 )
- Hydrochloric acid 37% (Merck, catalog number: 100317 )
- PCR mastermix (2x) (Thermo Fisher Scientific, catalog number: K0171 )
- PCR grade DMSO (Sigma-Aldrich, catalog number: D9170 )
- Tryptone (HiMedia Laboratories, catalog number: RM014 )
- Yeast extract powder (HiMedia Laboratories, catalog number: RM027 )
- LB growth media (see Recipes)
- Carbonate buffer (see Recipes)
- Tris-HCl binding buffer (see Recipes)
- Glycine-HCl elution buffer (see Recipes)
- Tris neutralization solution (see Recipes)
- Phenylboronic acid (PBA) coating solution (see Recipes)
- Phosphate buffered saline (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, model: FinnpipetteTM F1 variable volume single channel pipettes)
- Incubator-Shaker (Eppendorf, model: Innova® 44 )
- 4 °C refrigerator (Vestfrost Solutions, model: AKG 377 )
- -20 °C freezer (Vestfrost Solutions, model: CFS 344 )
- pH meter (Hanna Instruments, model: pH 211 )
- Spectrophotometer (GE Healthcare, model: NanoVue Plus )
- Centrifuge (Eppendorf, model: 5430 R )
- Thermocycler (Bio-Rad Laboratories, model: C1000 TouchTM )
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Shorie, M. and Kaur, H. (2018). Microtitre Plate Based Cell-SELEX Method. Bio-protocol 8(20): e3051. DOI: 10.21769/BioProtoc.3051.
分类
分子生物学 > DNA > DNA-蛋白质相互作用
微生物学 > 微生物蛋白质组学 > 膜蛋白
生物物理学 > 生物工程 > 人工受体
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link