- EN - English
- CN - 中文
Extraction and Quantification of Polyphosphate (polyP) from Gram-negative Bacteria
从革兰氏阴性菌中提取和测定多聚磷酸盐
(*contributed equally to this work) 发布: 2018年09月20日第8卷第18期 DOI: 10.21769/BioProtoc.3011 浏览次数: 7032
评审: Valentine V TrotterAnonymous reviewer(s)

相关实验方案
酸水解-高效液相色谱法测定集胞藻PCC 6803中聚3-羟基丁酸酯的含量
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
2023年08月20日 1790 阅读

一种利用 TMSD 甲基化结合 LC-MS 分析法快速定量细胞培养物中长链聚异戊烯类磷酸盐的新方法
Dipali Kale [...] Britta Brügger
2023年11月20日 1549 阅读
基于高效液相色谱法的史氏分枝杆菌DisA环二腺苷酸(C-di-AMP)合成酶活性研究
Avisek Mahapa [...] Dipankar Chatterji
2024年12月20日 1761 阅读
Abstract
Polyphosphate (polyP), a universally conserved biomolecule, is composed of up to 1,000 phosphate monomers linked via phosphoanhydride bonds. Reaching levels in bacteria that are in the high nmoles per mg protein range, polyP plays important roles in biofilm formation and colonization, general stress protection and virulence. Various protocols for the detection of polyP in bacteria have been reported. These methods primarily differ in the ways that polyP is extracted and/or detected. Here, we report an improved method, in which we combine polyP extraction via binding to glassmilk with a very sensitive PolyP kinase/luciferase-based detection system. By using this procedure, we significantly enhanced the sensitivity of polyP detection, making it potentially applicable for mammalian tissues.
Keywords: Polyphosphate (多聚磷酸盐)Background
Polyphosphate (polyP), a biopolymer composed of linear chains of up to 1,000 inorganic phosphate monomers, is found in cells of all three domains of life. Yet, bacteria are the only organisms for which the enzymes of polyP metabolism have been well studied. Bacterial polyP kinase (PPK), which converts ATP into polyP, catalyzes both forward and reverse reactions. While synthesis of polyP is clearly the favored reaction in the cell, by providing sufficient amounts of ADP in vitro, the enzyme can be used to generate ATP from polyP, making a luciferase-based ATP detection possible (Ault-Riché et al., 1998). Bacteria lacking PPK are defective in biofilm formation, motility, persistence, and various stress responses, and show significantly increased sensitivity towards hypohalous acids (i.e., bleach) stress or phosphate starvation (Figure 1) (Rao et al., 2009; Gray et al., 2014; Maisonneuve and Gerdes, 2014; Gray and Jakob, 2015; Groitl et al., 2017).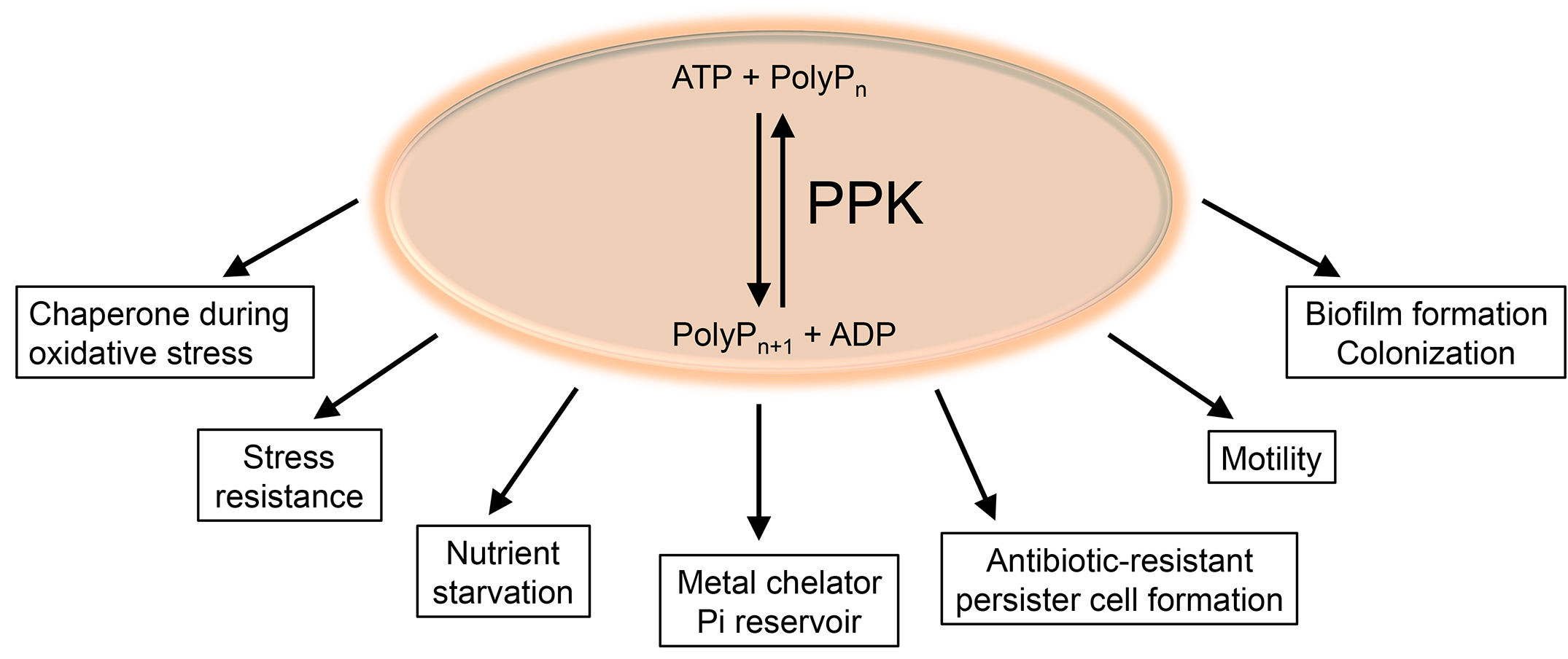
Figure 1. Synthesis of polyP and its role in Gram-negative Bacteria. The bacteria-specific polyphosphate kinase (PPK) reversibly catalyzes the conversion from ATP into polyP and ADP. Various functions for polyP have been described in Gram-negative bacteria, including its involvement in biofilm formation, colonization, motility, and formation of antibiotic-resistant persister cells. PolyP also contributes to the resistance of bacteria towards various stresses, including oxidative stress and starvation, and serves as metal chelator and Pi reservoir.
Given the many roles that polyP plays in Gram-negative bacteria, PPK became attractive as drug target to interfere with biofilm formation, make bacteria less persistent, and sensitize them towards physiological oxidants such as bleach (Dahl et al., 2017). Therefore, reliable and sensitive methods to determine the polyP levels in vivo are necessary. Several methods for the extraction and detection of polyP have been reported in Bio-protocol, including extraction of polyP with (i) perchloric acid, (ii) sodium hypochlorite, and (iii) phenol/chloroform and detection of polyP via visualization with urea-PAGE or colorimetric assays using malachite green or molybdenum blue (Gomez Garcia, 2014; Canadell et al., 2016; Ota and Kawano, 2017). In this protocol, we combined extraction of polyP via binding to glassmilk (Ault-Riché et al., 1998) with a very sensitive two-step enzyme-based detection system. First, the extracted polyP is converted into ATP by E. coli PPK in the presence of ultra-pure ADP. The ATP levels are then quantified using a luciferase-based detection system and corrected for cellular ATP. In comparison to the urea-PAGE or the colorimetric methods, the luciferase-based detection allows the quantification of much lower levels of polyP. This protocol has been successfully applied to quantify polyP levels from Pseudomonas aeruginosa.
Materials and Reagents
- Microcentrifuge tubes, 1.5 ml, clear (BioExpress, catalog number: C-3260-1 )
- Silica membrane spin columns (Epoch Life Science, EconoSpinTM, catalog number: 1920-250 )
- TempPlate non-skirted 96-well PCR plate, low profile, natural (USA Scientific, catalog number: 1402-9500 )
- 96-well plate solid white (Corning, catalog number: 3912 )
- 96-well plate clear flat bottom (Corning, catalog number: 3596 )
- Bradford reagent (Bio-Rad Laboratories, catalog number: 5000006 )
- Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A3059-100G )
- Tris (Fisher Scientific, catalog number: BP152-5 )
- Ultra-pure ADP (Cell Technology, catalog number: ADP100-2 )
- E. coli polyphosphate kinase (PPK) (for expression and purification of E. coli PPK, see Gray et al., 2014)
- Dithiothreitol (DTT), > 99% pure, protease-free (Gold Biotechnology, catalog number: DTT25 )
- Sodium-dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771-1KG )
- 95% v/v Ethanol
- QuantiLum® Recombinant Luciferase (Promega, catalog number: E1701 ; exact concentration of aliquots may vary)
Notes:- Prepare aliquots of the enzyme and store in small glass vials at -80 °C.
- Thaw and use the enzyme for each experiment.
- Discard all unused product.
- Do not subject to freeze-thaw cycles.
- Guanidine thiocyanate (Sigma-Aldrich, catalog number: G6639-250G )
- Silicon dioxide (Sigma-Aldrich, catalog number: S5631 )
- NaCl (Fisher Scientific, catalog number: S271-10 )
- EDTA (Fisher Scientific, catalog number: BP120-500 )
- HEPES (pH 7.5) (Fisher Scientific, catalog number: BP310-100 )
- Ammonium sulfate (Fisher Scientific, catalog number: A702-500 )
- Tricine buffer (pH 7.8) (Sigma-Aldrich, catalog number: T0377 )
- MgSO4 (Thermo Fisher Scientific, catalog number: M63500 )
- Sodium azide (MP Biomedicals, catalog number: 210289125 )
- Luciferin (Biotium, catalog number: 10101-1 )
- Glycylglycine (pH 7.8) (MP Biomedicals, catalog number: 210185610 )
- ddH2O
- Hydrochloric acid (Fisher Scientific, catalog number: A144212 )
- GITC lysis buffer (see Recipes)
- Glassmilk (see Recipes)
- New Wash (NW-) Buffer (see Recipes)
- Elution Buffer (see Recipes)
- PPK buffer (see Recipes)
- Luciferase reaction buffer (see Recipes)
- Luciferin working solution (see Recipes)
Equipment
- Eppendorf Pipettes: 100-1,000 μl, 20-200 μl, 2-20 μl, 1-10 μl
- Thermomixer (Eppendorf, model: 5350 )
- Microcentrifuge (Eppendorf, model: 5415D )
- Incubator 37 °C (VWR, model: 1555 )
- -80 °C Freezer (Eppendorf, New BrunswickTM, model: Innova® U725 , catalog number: U9440-0002)
- Fluostar Omega Plate Reader with luminescence reading function and injector module (BMG Labtech, catalog number: 0415-102 )
- pH meter
- Vortexer
- Refrigerator (4 °C)
Software
- Fluostar Omega Control and Evaluation Software (BMG Labtech, catalog number: 1300-501)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Dahl, J. U., Xie, L. and Jakob, U. (2018). Extraction and Quantification of Polyphosphate (polyP) from Gram-negative Bacteria. Bio-protocol 8(18): e3011. DOI: 10.21769/BioProtoc.3011.
分类
微生物学 > 微生物生物化学 > 其它化合物
微生物学 > 微生物生理学 > 胁迫反应
生物化学 > 其它化合物 > 多磷酸盐
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link