- EN - English
- CN - 中文
Enzymatic Synthesis and Fractionation of Fluorescent PolyU RNAs
荧光polyU RNAs的酶促合成及分离
发布: 2018年09月05日第8卷第17期 DOI: 10.21769/BioProtoc.2988 浏览次数: 5237
评审: Vamseedhar RayaproluKarthik KrishnamurthyOmar Akil
Abstract
The physical properties of viral-length polyuridine (PolyU) RNAs, which cannot base-pair and form secondary structures, are compared with those of normal-composition RNAs, composed of comparable numbers of each of A, U, G and C nucleobases. In this protocol, we describe how to synthesize fluorescent polyU RNAs using the enzyme polynucleotide phosphorylase (PNPase) from Uridine diphosphate (UDP) monomers and how to fractionate the polydisperse synthesis mixture using gel electrophoresis, and, after electroelution, how to quantify the amount of polyU recovered with UV-Vis spectrophotometry. Dynamic light scattering was used to determine the hydrodynamic radii of normal-composition RNAs as compared to polyU. It showed that long polyU RNAs behave like linear polymers for which the radii scale with chain length as N1/2, as opposed to normal-composition RNAs that act as compact, branched RNAs for which the radii scale as N1/3.
Keywords: Homopolymer polyU RNA (均聚物 polyU RNA)Background
PolyU as a Physical Object: PolyU is an abiological RNA molecule composed of repeated uridine residues, and therefore, it cannot Watson-Crick base-pair and is devoid of RNA secondary structure (Martin and Ames, 1962; Richards et al., 1963). PolyU has very weak base-stacking energy resulting in a lack of helical ordering as well–except below 4 °C (Richards et al., 1963). Due to this lack of structure polyU RNA can be described physically as a random coil (Inners and Felsenfeld, 1970), as opposed to normal RNA molecules that form complicated secondary/tertiary structures, making them more compact physical objects (Yoffe et al., 2008; Fang et al., 2011; Gopal et al., 2012; Garmann et al., 2015).
We therefore expect polyU to be less compact than viral RNA of the same length. Indeed dynamic light scattering (DLS) measurements (see Figure 3) indicate that in RNA assembly buffer (see Recipes) at 25 °C, polyU molecules are about 25% larger in average diameter than branched RNAs of the same length. Additionally, we expect polyU to be more flexible than normal-composition RNAs, as it lacks secondary and tertiary structure, resulting in a more extended, flexible molecule.
Synthesis and Fractionation of Long PolyU RNAs: The synthesis protocol used herein was developed from existing protocols used by Vanzi et al. (2003) and van den Hout et al. (2011). Specifically, polyU RNAs were synthesized from Uridine diphosphate (UDP) monomers using the enzyme polynucleotide phosphorylase (PNPase), which indiscriminately adds nucleotide diphosphates (NDPs) to the 3' end of oligonucleotides, in this case a 20-nt-long polyU seed oligo with a 5' cy5 fluorescent label (Beren et al., 2017). It is important to note that the oligo sequence and length can be changed as desired, and it can be tagged at the 5' end with fluorophores, linkers, or modified bases, enabling the synthesis of polyU with customizable markers. Because of its lack of base-pairing, polyU cannot be visualized in electrophoresis gels by the usual intercalating nucleic acid stains. For this reason, we chose to utilize a fluorophore at the 5' end of the RNA. Additionally, it is possible to make polyA, polyG or polyC using the same PNPase, by simply adding ADP, GDP or CDP, respectively (Grunberg-Manago et al., 1956).
The synthesis reactions produce a polydisperse mixture of fluorescent polyU RNAs ranging in length from 200 to 10,000 nts, with shorter lengths being represented in significantly higher copy number as determined by fluorescence detection in denaturing agarose gel electrophoresis (see Figure 1). After running the RNA mixture alongside a denatured ssRNA ladder (see Figure 1), it was fractionated by electroeluting excised portions of the gel using the ladder as a reference. In fact, we often run many lanes of polyU in parallel to increase the amount of fractionated polyU produced from a given gel, as we usually recover between 30% and 60% of the RNA after gel electroelution.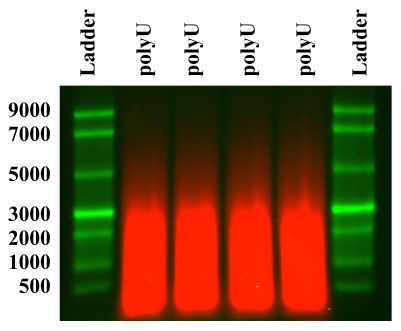
Figure 1. Denaturing electrophoresis gel analysis of unfractionated polyU. Electrophoresis gel analysis of unfractionated polyU (red, cy5 fluorescence) after synthesis using PNPase, compared with a single-stranded (ss) RNA ladder (green, ethidium bromide nucleic acid stain). This gel illustrated the high amount of polydispersity in the polyU generated from the synthesis reaction. The brightest band in the RNA ladder is 3,000-nt-long ssRNA.
We used this process to fractionate polyU into the following length fractions: 500-1,500, 1,500-2,500, 2,500-3,500, 3,500-5,000, 5,000-7,000 and 7,000-9,000 nt. The amount of polyU purified was quantified using UV-Vis absorbance measurements, and the absorbance ratio at 260 nm/280 nm was used as a measure of RNA purity (A260/A280 greater than 2.0). These fractionated polyU samples were then examined on a denaturing agarose gel to demonstrate that single contiguous RNA bands were obtained (see Figure 2).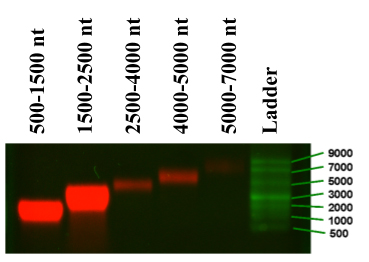
Figure 2. Denaturing electrophoresis gel analysis of fractionated polyU. From right to left: ssRNA ladder, 5,000-7,000, 4,000-5,000, 2,500-4,000, 1,500-2,500 and 500-1,500 nt polyU RNAs. The decrease in band intensity for higher-molecular-weight polyU molecules is due to the fact that an equal mass of polyU was loaded per lane, resulting in many fewer RNAs in the higher-molecular-weight fractions, and subsequently, a decreased fluorescence signal (because of there being only one label per RNA molecule). (Adapted from Beren et al., 2017)
Materials and Reagents
- PolyU synthesis
- 100 kDa Amicon filters (Merck, catalog number: UFC510096 )
- 6-8 kDa MW cutoff dialysis tubing (Repligen, Spectra/Por®, catalog number: 132650 )
- Eppendorf tubes (Eppendorf, catalog number: 022363204 )
Note: We do not utilize RNase-free tubes, but always autoclave the Eppendorf tubes before use. - RNase Inhibitor (Roche Diagnostics, catalog number: 03 335 399 001 , stored at -20 °C)
- Polynucleotide phosphorylase (PNPase) from Synechocystis sp. (Sigma-Aldrich, catalog number: N9914-100UG , stored at -80 °C)
- Uridine diphosphate (UDP, Sigma-Aldrich, catalog number: 94330 , stored at -20 °C)
- 10-25 nt RNA seed oligo (IDT, DNA, stored in TE buffer at -80 °C)
Note: We utilized a 20-nt-long polyU RNA seed oligo for our experiments. - Tris base (Merck, catalog number: 648310-2.5KG ), for making buffer Tris pH 9
- EDTA (Merck, Omnipur®, catalog number: 4050-1KG )
- Magnesium chloride hexahydrate (MgCl2•6H2O) (Merck, Omnipur®, catalog number: 5980-500GM )
- Hydrochloric Acid (Thermo Fisher Scientific, catalog number: FLA144500 )
- Synthesis buffer (stored at 4 °C) (see Recipes)
- TE buffer (stored at 4 °C) (see Recipes)
- PolyU fractionation
- 100 kDa Amicon filters (Merck, catalog number: UFC510096 )
- 6-8 kDa MW cutoff dialysis tubing (Repligen, Spectra/Por®, catalog number: 132650 )
- Eppendorf tubes (Eppendorf, catalog number: 022363204 )
Note: We do not utilize RNase-free tubes, but always autoclave the Eppendorf tubes before use. - Dialysis tubing clips (Repligen, Spectra/Por®, catalog numbers: 132743 and 132735 )
- Sodium bicarbonate (Merck, catalog number: SX0320-1 )
- RNase Inhibitor (Roche Diagnostics, catalog number: 03 335 399 001 , stored at -20 °C)
- Polynucleotide phosphorylase (PNPase) from Synechocystis sp. (Sigma-Aldrich, catalog number: N9914-100UG , stored at -80 °C)
- ssRNA ladder (New England Biolabs, catalog number: N0362S , stored at -80 °C)
- 1% agarose gel (prepared in TAE buffer, agarose is purchased from Merck, Omnipur®, Calbiochem, catalog number: 2125-500GM )
- Formamide (Acros organics, catalog number: 327235000 , stored at 4 °C)
- Gel red (Biotium, catalog number: 41001 )
- Tris base (Merck, catalog number: 648310-2.5KG ), for making buffer Tris pH 8
- Acetic acid (Sigma-Aldrich, catalog number: 695092 )
- EDTA (Merck, Omnipur®, catalog number: 4050-1KG )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S671-10 )
- Potassium chloride (KCl) (Merck, catalog number: 7300-500GM )
- Magnesium chloride hexahydrate (MgCl2•6H2O) (Merck, Omnipur®, catalog number: 5980-500GM )
- Hydrochloric Acid (Thermo Fisher Scientific, catalog number: FLA144500 )
- TAE buffer (stored at 4 °C) (see Recipes)
- RNA assembly buffer (see Recipes)
Equipment
Note: Everything is sterilized before use, either by autoclaving or rinsing with 70% ethanol.
- Scalpel (Surgical Design, model: SCALPEL#15 )
- 70% Ethanol (We dilute 100% ethanol with ddH2O) (Decon Labs, catalog number: V1001 )
- Micropipette volumes used range from 1-1,000 μl (Mettler-Toledo, Rainin, model: Pipet-Lite XLS LTS )
- Micropipette tips: volumes used range from 1-1,000 μl (filtered, sterile aerosol tips with high recovery for Rainin LTS Pipettemen; VWR, catalog numbers: 83009-688 , 82003-196 , 82003-198 )
- Autoclave
- Heat block (set to 70 °C) (Fisher Scientific, catalog number: 11-718-2Q )
- Ice or a cold block for RNA work (VWR, catalog number: 414004-285 )
- 37 °C incubator (Precision, catalog number: 51221089 )
- Centrifuge that can run to 5,000 x g (Eppendorf, model: 5804 R )
- Gel electrophoresis equipment
Gel tank/box combo (Fisher Scientific, catalog number: FB-SB-710 )
Gel combs (Thermo Fisher Scientific, catalog numbers: B1A-10 and B1A-6 )
Power supply (Fisher Scientific, catalog number: FB1000Q ) - Laminar flow hood, for RNA work
- UV lamp, for imaging the ssRNA ladder in the gel (Spectronics, model: ENF-260C )
- Nanodrop, UV-Vis spectrophotometer for measuring RNA concentrations
- Malvern particle sizer Nano-ZS
- Milli-Q UV Plus (Merck, catalog number: ZD60115UV )
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Beren, C., Liu, K. N., Dreesens, L. L., Knobler, C. M. and Gelbart, W. M. (2018). Enzymatic Synthesis and Fractionation of Fluorescent PolyU RNAs. Bio-protocol 8(17): e2988. DOI: 10.21769/BioProtoc.2988.
分类
分子生物学 > RNA > RNA 标记
生物化学 > RNA > RNA结构
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link