- EN - English
- CN - 中文
Single and Multiplexed Gene Editing in Ustilago maydis Using CRISPR-Cas9
使用CRISPR-Cas9对玉米黑粉菌进行单个和多个基因的编辑
发布: 2018年07月20日第8卷第14期 DOI: 10.21769/BioProtoc.2928 浏览次数: 9922
评审: Aksiniya AsenovaShahin S. AliAnonymous reviewer(s)
Abstract
The smut fungus Ustilago maydis is an established model organism for elucidating how biotrophic pathogens colonize plants and how gene families contribute to virulence. Here we describe a step by step protocol for the generation of CRISPR plasmids for single and multiplexed gene editing in U. maydis. Furthermore, we describe the necessary steps required for generating edited clonal populations, losing the Cas9 containing plasmid, and for selecting the desired clones.
Keywords: CRISPR-Cas9 (CRISPR-Cas9)Background
The basidiomycete fungus U. maydis is a model organism that has enabled important discoveries in topics like DNA repair and homologous recombination, mating, sexual development, secondary metabolism, filamentous growth, RNA transport and virulence (Holliday, 2004; Steinberg, 2007; Bakkeren et al., 2008; Holloman et al., 2008; Lanver et al., 2017; Niessing et al., 2018). U. maydis is a biotrophic plant pathogen that causes smut disease on maize. As a typical member of the large group of smut fungi its sexual development is intimately coupled with its ability to colonize plants. The popularity of U. maydis as a model organism resides in its short pathogenic life cycle that is completed in less than two weeks, its accessibility for forward and reverse genetics that includes the establishment of self-replicating plasmids (Tsukuda et al., 1988), the development of solopathogenic strains able to cause disease without mating and an available well-annotated genome (Kämper et al., 2006). In U. maydis, homologous recombination-based genome manipulations are very efficient, but become tedious when several genes have to be modified because the number of selectable markers is limited (Schuster et al., 2016). The most recent addition to the technical toolboxes for this organism has been the adaptation of the CRISPR-Cas9 genome editing technology (Schuster et al., 2016) and its development for multiplexed editing (Schuster et al., 2018). The CRISPR-Cas9 genome editing technology, developed from research in bacterial immune systems (Barrangou et al., 2007), has revolutionized molecular biology. The system allows efficient modification of almost any given sequence (Jinek et al., 2012) and has been established for a large number of organisms including filamentous fungi (Deng et al., 2017; Shi et al., 2017).
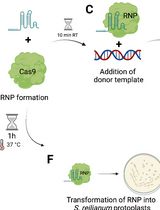
The CRISPR-Cas9 technology established for U. maydis is based on an all-in-one plasmid approach. A self-replicating plasmid (Tsukuda et al., 1988) is used as a backbone to provide the U. maydis codon-optimized Cas9 gene and the U6 promoter for sgRNA expression. This plasmid is unstable without selection and is rapidly lost (Figure 1 and Schuster et al. [2016]). The system proved very efficient for disruption of single genes reaching on average 70% of edited cells in progeny of one transformant (Schuster et al., 2016). A multiplexing version of the system was generated by using tRNA promoter-based expression cassettes that enabled the expression of several sgRNAs from the same plasmid (Schuster et al., 2018). This modification combined with elevated and longer expression of the Cas9 protein was used for the simultaneous disruption of five effector genes with 70% efficiency (Schuster et al., 2018). The multiplexed version of CRISPR-Cas9 has also been successfully applied for elucidating the contribution of oligopeptide transporters to virulence (Lanver et al., 2018).
Materials and Reagents
- Consumables
- Sterile pipette tip
- Toothpick
- 60 mm x 15 mm round Petri dishes
- Sterile pipette tip
- Competent cells
E. coli competent cells e.g., One Shot TOP10 Chemically Competent E. coli (Thermo Fisher Scientific, catalog number: C404010 ) - Plasmids and double-stranded DNA
- Oligonucleotides 10 pmol/µl
- oMS59: 5’ ATTCGTGATTTACACCAAACACGC 3’
- oMS49: 5’ CCCCTCGTCTCGCGCCTCATTGGTCGAATTG 3’
- oSR224: 5’ CTACACTCAGCACACGATGT 3’
- oMS59: 5’ ATTCGTGATTTACACCAAACACGC 3’
- Enzymes and buffers
- Kits
- Antibiotics
- Reagents
- Tryptone (BD Biosciences)
- Yeast extract (BD Biosciences)
- Sodium chloride (NaCl)
- Peptone (BD Biosciences)
- Sucrose (BD Biosciences)
- Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Bacto agar (BD Biosciences)
- Potato Dextrose Broth (BD Biosciences)
- Tryptone (BD Biosciences)
- Media (see Recipes)
- Yeast extract tryptone (dYT) liquid medium + 100 µg/ml ampicillin
- YT agar plates + 100 µg/ml ampicillin
- YEPS light liquid medium +/- 2 µg/ml carboxin
- Two layered regeneration agar light (RegAgar) plates (Bottom layer (10 ml) with 4 µg/ml carboxin and top layer (10 ml) without carboxin (Prepare fresh))
- Potato dextrose (PD) agar plates +/- 2 µg/ml carboxin
- Yeast extract tryptone (dYT) liquid medium + 100 µg/ml ampicillin
Equipment
Note: No equipment from specific manufacturers is required. Any equivalent device can be used.
- Pipettes
- Computer with internet access
- Plate incubators (28 °C, 37 °C)
- Shakers (28 °C, 37 °C)
- ThermoMixer (e.g., Eppendorf, model: ThermoMixer® C , catalog number: 5382000015)
- Thermocycler (e.g., Analytik, Jena, Biometra, catalog number: 846-2-070-301 )
- Tabletop centrifuge (e.g., Thermo Fisher Scientific, catalog number: 75008801 )
Software
No specific software is required. Any appropriate alignment program like CloneManager 9 Professional Edition (Scientific & Educational Software) or CLC Main Workbench (QIAGEN) can be used.
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Schuster, M., Trippel, C., Happel, P., Lanver, D., Reißmann, S. and Kahmann, R. (2018). Single and Multiplexed Gene Editing in Ustilago maydis Using CRISPR-Cas9. Bio-protocol 8(14): e2928. DOI: 10.21769/BioProtoc.2928.
分类
微生物学 > 微生物遗传学 > 基因组编辑
分子生物学 > DNA > DNA 修饰
微生物学 > 微生物遗传学 > CRISPR-Cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link