- EN - English
- CN - 中文
Implementation of Blue Light Switchable Bacterial Adhesion for Design of Biofilms
通过蓝光控制的细菌粘附进行生物膜设计
发布: 2018年06月20日第8卷第12期 DOI: 10.21769/BioProtoc.2893 浏览次数: 7173
评审: Elizabeth LibbyTimo A LehtiKumari Sonal Choudhary

相关实验方案

从沙门氏菌鼠伤寒血清中纯化细菌淀粉样蛋白“Curli”并检测受感染宿主组织中的 Curli
Murugesan Sivaranjani [...] Aaron P. White
2022年05月20日 3236 阅读
Abstract
Control of bacterial adhesions to a substrate with high precision in space and time is important to form a well-defined biofilm. Here, we present a method to engineer bacteria such that they adhere specifically to substrates under blue light through the photoswitchable proteins nMag and pMag. This provides exquisite spatiotemporal remote control over these interactions. The engineered bacteria express pMag protein on the surface so that they can adhere to substrates with nMag protein immobilization under blue light, and reversibly detach in the dark. This process can be repeatedly turned on and off. In addition, the bacterial adhesion property can be adjusted by expressing different pMag proteins on the bacterial surface and altering light intensity. This protocol provides light switchable, reversible and tunable control of bacteria adhesion with high spatial and temporal resolution, which enables us to pattern bacteria on substrates with great flexibility.
Keywords: Bacterial adhesion (细菌粘附)Background
Controlling the biofilm formation is crucial to understand the social interactions between bacteria in naturally occurring biofilm (Flemming et al., 2016). This is also particularly important for the biotechnological application of biofilms in biocatalysis, biosensing and waste treatment (Zhou et al., 2013; Jensen et al., 2016). The biofilm formation always begins with the bacterial adhesion to a substrate, which determines the spatial organization in biofilms (Liu et al., 2016; Nadell et al., 2016). Many strategies have been proposed to control bacterial adhesion such as modifying bacterial surface with bio-orthogonal reactive groups via liposome fusion (Elahipanah et al., 2016), immobilization of adhesive molecules on the substrates (Sankaran et al., 2015; Zhang et al., 2016; Peschke et al., 2017) and conjugating surface tags on bacteria (Poortinga et al., 2000; Rozhok et al., 2005; Lui et al., 2013). Among these, the light responsive approaches provide the highest spatiotemporal control, which is important to precisely control the fine structure of the biofilms. For instance, azobenzene linkers have been used as a photoswitchable tool to reversibly control the bacterial adhesion to substrates by altering the presentation of mannose, which is recognized by the bacterial surface receptor FimH (Voskuhl et al., 2014; Weber et al., 2014; Sankaran et al., 2015). In addition, azobenzene-based molecules have also been used to control bacteria adhesion to mammalian cells (Mockl et al., 2016), bacterial quorum sensing (Van der Berg et al., 2015) and biofilm formation (Hu et al., 2016) with UV-light. One of the major drawbacks of using UV-light is that it is toxic to bacteria. In this protocol, we present a new approach of how to control bacterial adhesion to substrates with blue light based on photoswitchable proteins. Besides being a non-invasive, reversible and tuneable technique to control bacterial adhesion to substrates, it also provides high spatiotemporal control required to form well-defined biofilms. Photoswitchable proteins are commonly used in the field of optogenetics to regulate gene expression, receptor activation and protein localization in cells with visible light (Müller and Weber, 2013; Tischer and Weiner, 2014). These optogenetic systems are very sensitive to visible light, bioorthogonal and noninvasive. Furthermore, these proteins are genetically encoded so they can be sustainably expressed in the cell. Here, we used the blue light responsive proteins, nMag and pMag, as photoswitches control bacterial adhesion. These proteins heterodimerize under blue light (480 nm) and dissociate from each other in the dark (Kawano et al., 2015). The strength and back conversion kinetics of the nMag and pMag interaction are different for the point mutants. The point mutant pMagHigh (and nMagHigh) has a stronger interaction with its binding partners and slower back conversion, while the opposite is true for the mutant pMagFast1 (and nMagFast1) (Zoltowski et al., 2009).
In our method we display the first interaction partner of the photoswitchable proteins, pMagHigh, pMag or pMagFast1 on the surface of E. coli using the circularly permutated OmpX (outer membrane protein X) protein (Daugherty, 2007). The pMag variants are attached through their C-terminal to the OmpX protein. The second interaction partner the photoswitchable protein, nMagHigh, is immobilized through a His6-tag at its C-terminal on a glass substrate with a PEG (polyethylene glycol) coating, which contains a Ni2+-NTA group (Schenk et al., 2014). This setup allows bacteria expressing pMag proteins on their surfaces to adhere to nMagHigh functionalized substrates under blue light when the two proteins interact but not in the dark. (Figure 1)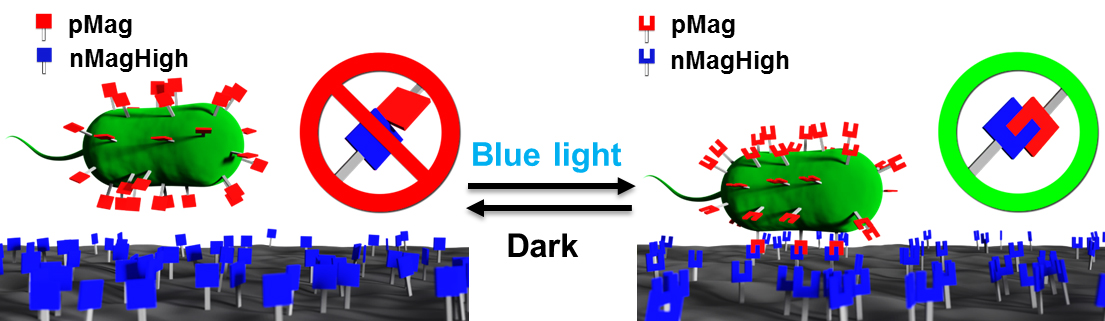
Figure 1. The engineered E. coli that express pMag proteins on their surface adhere to nMagHigh modified substrates under blue light. In the dark, the pMag-nMag interaction is reversed, which leads to the detachment of the bacteria from the substrate. Reproduced with permission from Chen and Wegner (2017).
Materials and Reagents
- Pipette tips (STARLAB, catalog number: S1111-6700 )
- 50 ml Falcon tube (Greiner Bio One International, catalog number: 227261 )
- 1.5 ml Eppendorf tube (Eppendorf, catalog number: 0030120086 )
- 0.2 ml PCR tubes (Thermo Fisher Scientific, catalog number: AB0620 )
- 0.45 µm cellulose filter (Carl Roth, catalog number: KH55.1 )
- Ni-NTA column (GE Healthcare, catalog number: 17524801 )
- 50 ml syringe (VWR, catalog number: 53548-010)
Manufacturer: Air-Tite Products, catalog number: 4850001000 . - 20 x 20 mm glass slides (VWR, catalog number: 631-0122 )
Notes: The glass slide is used as the glass surface for the protein functionalization and bacterial experiments. - Parafilm (Sigma-Aldrich, Bemis, catalog number: P7668 )
- Aluminum foil (Carl Roth, catalog number: 1770.1 )
- 35 mm Petri dish (SARSTEDT, catalog number: 82.1135.500 )
- Dialysis tubing (Repligen, Spectrum, catalog number: 132592 )
- Plasmid pB33eCPX (Addgene plasmid) (Addgene, catalog number: 23336 )
- GFP and mCherry pTrc99A plasmids (Prof. Victor Sourjik lab, Chen and Wegner, 2017)
- nMagHigh pET-21b(+) plasmid (Genescript, Chen and Wegner, 2017)
- nMagHigh-eCPX plasmid (homemade, Chen and Wegner, 2017)
Note: The nMagHigh gene is inserted between the KpnI and SacI cutting sites of pB33eCPX. - pMag-eCPX, pMagHigh-eCPX, pMagFast1-eCPX plasmids (homemade, Chen and Wegner, 2017)
Note: The different pMag variants are generated by point mutagenesis from the nMagHigh-eCPX plasmid using QuikChange II Site-Directed Mutagenesis Kit. - E. coli K12 MG1655 (DSMZ, catalog number: 18039 )
- BL21(DE3) competent E. coli (homemade, Chen and Wegner, 2017)
- DH5α competent E. coli (homemade, Chen and Wegner, 2017)
- PBS Tablets (Thermo Fisher Scientific, GibcoTM, catalog number: 18912014 )
- Mowiol (Sigma-Aldrich, catalog number: 81381 )
- LB medium (Carl Roth, catalog number: X968.3 )
- Ampicillin (Carl Roth, catalog number: HP62.2 )
- IPTG (Sigma-Aldrich, catalog number: I6758 )
- PMSF (Sigma-Aldrich, catalog number: P7626 )
- DTT (Sigma-Aldrich, catalog number: D0632-10G )
- PEG-azide (homemade)
- Triethylamine (Sigma-Aldrich, catalog number: T0886 )
- Toluene, anhydrous (Alfa Aesar, catalog number: 41464-AK )
- EDTA (Sigma-Aldrich, catalog number: 798681 )
- NiCl2 (Sigma-Aldrich, catalog number: 339350 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- L-arabinose (Sigma-Aldrich, catalog number: A3256 )
- Paraformaldehyde, reagent grade (Sigma-Aldrich, catalog number: P6148 )
- Tris Base (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Imidazole (Sigma-Aldrich, catalog number: I2399 )
- L-Ascorbic acid (Sigma-Aldrich, catalog number: A5960 )
- NTA-alkyne (homemade, Schenk et al., 2014)
- PEG azide (homemade, Schenk et al., 2014)
- Copper sulfate (CuSO4) (Sigma-Aldrich, catalog number: 451657 )
- 30% H2O2 (Carl Roth, catalog number: 8070.4 )
- H2SO4 (Sigma-Aldrich, catalog number: 30743)
- TEM (Transmission electron microscopy) grid (Ted Pella, catalog number: 1GC150 )
- Centrifuge tubes 500 ml and 50 ml (Thermo Fisher Scientific, catalog numbers: 3141-0500 , 3119-0050 )
- Methanol (Fisher Scientific, catalog number: 10224490 )
- N2 gas (Westfalen)
- Ethyl acetate (VWR, catalog number: 23882.321 )
- Picodent twinsil 22 (Picodent, catalog number: 13001000 )
- Buffer A (see Recipes)
- Buffer B (see Recipes)
- Click reaction solution (see Recipes)
- Riranha solution (see Recipes)
Equipment
- Dumont #7 Tweezers (Carl Roth, catalog number: K344.1 )
Note: Dumont #7 Tweezers is used to pick up the glass slides. - 0.1-2.5 μl, 0.5-10 μl, 10-100 μl, 100-1,000 μl Pipettes (Eppendorf, catalog numbers: 3123000012 , 3123000020 , 3123000047 , 3123000063 )
- Vortexer (neoLab, catalog number: 7-0092 )
- Microcentrifuge (VWR, model: Micro Star 17, catalog number: 521-1646 )
- High-speed centrifuge (Beckman Coulter, model: Avanti® J-26S )
- Rotors for high-speed centrifuge (Beckman Coulter, models: JA-10 , JA-25.50 )
- Incubator (VWR, catalog number: 444-0732 )
- Sonicator (OMNI, model: Sonic Ruptor 400 )
- Invert fluorescence microscope (Leica Microsystems, model: Leica DMi8 )
- Ultrasonic cleaner (BANDELIN electronic, model: Sonorex Super RK 31 )
- Blue LED panel (Albrillo, model: LL-GL003 )
- OD Meter (Biochrom, BioWave, model: WPA CO8000 )
- Nanodrop (Thermo Fisher Scientific, model: NanoDropTM 8000 )
Software
- ImageJ
- Originlab
Note: Oringinlab is used for data analysis.
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Chen, F. and Wegner, S. V. (2018). Implementation of Blue Light Switchable Bacterial Adhesion for Design of Biofilms. Bio-protocol 8(12): e2893. DOI: 10.21769/BioProtoc.2893.
分类
微生物学 > 微生物生物膜 > 生物膜培养
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











