- EN - English
- CN - 中文
The Long-lived Protein Degradation Assay: an Efficient Method for Quantitative Determination of the Autophagic Flux of Endogenous Proteins in Adherent Cell Lines
长寿蛋白降解测定法:定量测定粘附细胞系中内源性蛋白自噬通量的有效方法
发布: 2018年05月05日第8卷第9期 DOI: 10.21769/BioProtoc.2836 浏览次数: 11043
评审: Alessandro DidonnaPia GiovannelliManasi K. Mayekar
Abstract
Autophagy is a key player in the maintenance of cellular homeostasis in eukaryotes, and numerous diseases, including cancer and neurodegenerative disorders, are associated with alterations in autophagy. The interest for studying autophagy has grown intensely in the last two decades, and so has the arsenal of methods utilised to study this highly dynamic and complex process. Changes in the expression and/or localisation of autophagy-related proteins are frequently assessed by Western blot and various microscopy techniques. Such analyses may be indicative of alterations in autophagy-related processes and informative about the specific marker being investigated. However, since these proteins are part of the autophagic machinery, and not autophagic cargo, they cannot be used to draw conclusions regarding autophagic cargo flux. Here, we provide a protocol to quantitatively assess bulk autophagic flux by employing the long-lived protein degradation assay. Our procedure, which traces the degradation of 14C valine-labelled proteins, is simple and quick, allows for processing of a relatively large number of samples in parallel, and can in principle be used with any adherent cell line. Most importantly, it enables quantitative measurements of endogenous cargo flux through the autophagic pathway. As such, it is one of the gold standards for studying autophagic activity.
Keywords: Long-lived protein degradation (长寿蛋白降解)Background
Pulse-chase labelling approaches have been used to study protein turnover for decades. In the long-lived protein degradation (LLPD) assay described here, the proteome of cells in culture is radiolabelled with 14C valine and chased in order to follow the decline in radioactive proteins as readout of protein degradation. After an initial chase period, the cells are washed to eliminate the degradation products of short-lived proteins, which predominantly result from proteasomal activity. Thereafter, a second chase is initiated, and proper controls are included in order to monitor the autophagic degradation of long-lived proteins. We recently used this method to discover that the calcium-modulating compounds thapsigargin and A23187, which based on results with autophagic markers were previously widely believed to activate autophagy, actually completely block bulk autophagic flux (Engedal et al., 2013). The starting material used in that and previous protein degradation protocols was derived from cells grown in 6-well plates (Bauvy et al., 2009; Engedal et al., 2013) or more (Ronning et al., 1979; Seglen et al., 1979), or involved relatively high amounts of radioactivity (Mizushima et al., 2001; Fuertes et al., 2003a), which is expensive. Recently, we have downscaled and simplified the LLPD protocol to the validated time- and cost-efficient version that we present here.
An overview of the method is shown in Figure 1. To label proteins with radioactive valine, cells are seeded in 24-well plates in complete medium supplemented with 14C valine. As proteins are synthesised, they incorporate amino acids, including the 14C valine present in the medium, and therefore the amount of long-lived 14C valine-labelled proteins increases with time (Figure 1, first part of the curve). Valine is an optimal amino acid to use in the LLPD method, since it is a poorly metabolised amino acid, is well tolerated at high doses, and does not influence autophagy or protein degradation rates (Seglen et al., 1979). Moreover, free valine is readily exchanged over the plasma membrane (Seglen and Solheim, 1978), enabling efficient wash-out of released 14C valine. After 2-3 days of labelling, unincorporated 14C valine is removed by a simple wash procedure, and the cells receive new medium supplemented with a high concentration of non-radioactive (‘cold’) valine (‘chase medium’). The large surplus of cold valine prevents reincorporation of released 14C valine. Thus, from this point on the presence of free 14C valine is a result of endogenous protein degradation. After a chase period of 18 h, the free 14C valine that has been produced by degradation of short-lived proteins (predominantly due to proteasomal degradation) is washed out. Next, a second chase period, which we call the ‘sampling period’, is initiated along with experimental treatments and proper controls. Generally, we use a sampling period of 2-6 h to monitor the degradation of long-lived proteins. At the end of the sampling period, trichloroacetic acid (TCA) is added to precipitate intact proteins. The TCA-soluble fraction of degraded proteins (containing free amino acids and small peptides) is separated from the TCA-insoluble fraction (containing intact proteins) by centrifugation, and the radioactivity in each fraction is measured by liquid scintillation counting. This allows calculation of the rate of long-lived protein degradation in the sampling period, expressed as the percentage of radioactivity in the TCA-soluble fraction versus the total amount of radioactivity in the TCA-soluble and–insoluble fractions, divided by the duration of the sampling period (Figure 1).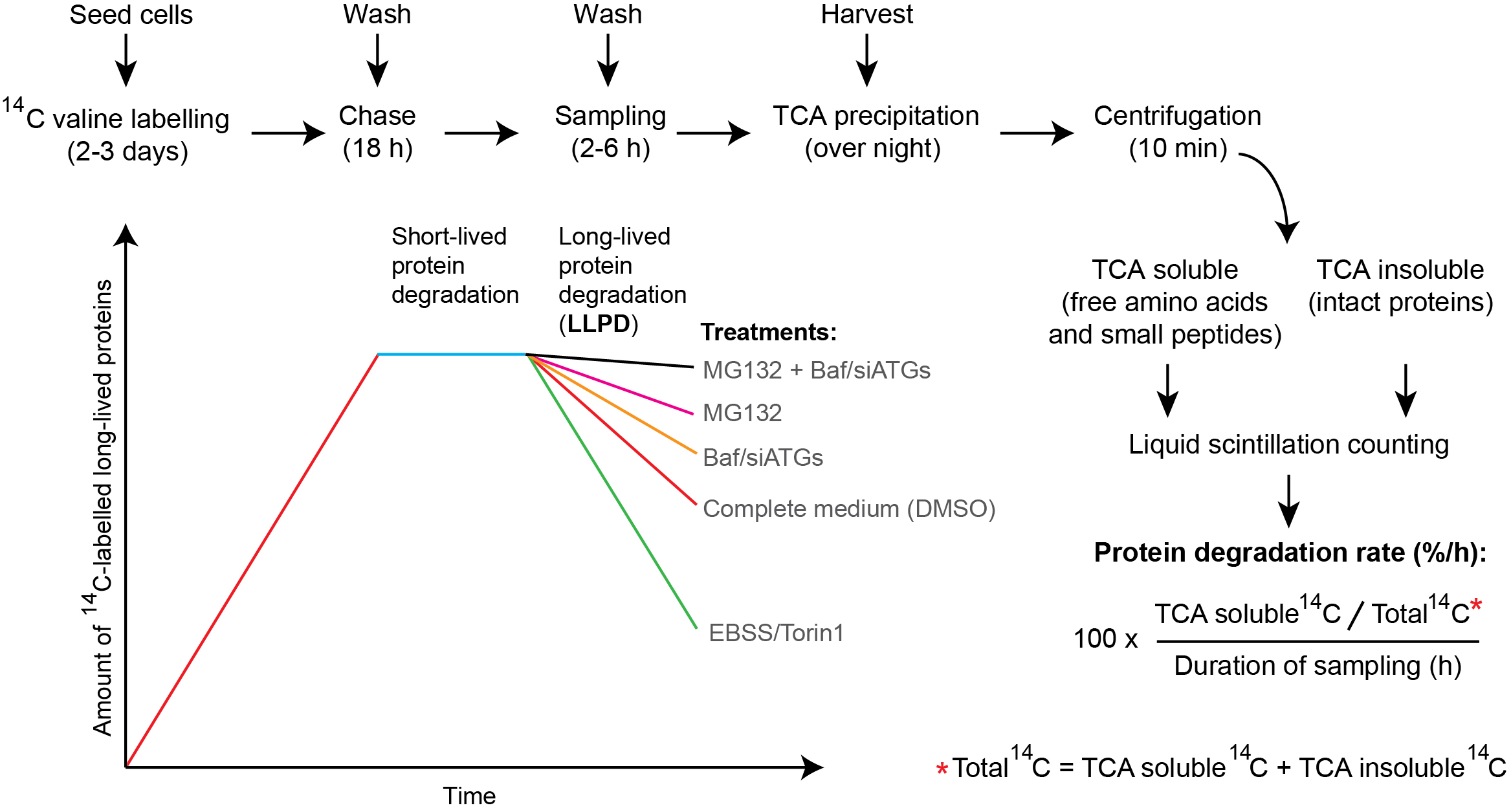
Figure 1. Overview of the long-lived protein degradation (LLPD) assay. During the labelling period (2-3 days), the amount of radioactive long-lived proteins increases with time. Thereafter, an 18 h chase period allows for degradation of the short-lived proteins and subsequent elimination of the released 14C valine by a washing step. Consequently, only the degradation of long-lived proteins is followed in the 2-6 h sampling period. Compared to cells kept in complete, nutrient-rich medium (red line), incubating cells with EBSS starvation medium or the mTOR-inhibitor Torin1 produces a very strong degradation of long-lived proteins in the sampling period, due to enhanced bulk autophagy (green line). Autophagic-lysosomal LLPD can be revealed by treatment with the lysosomal inhibitor Bafilomycin A1 (Baf) or RNAi-mediated silencing of key ATGs (siATGs) (yellow line), whereas the contribution to LLPD from the proteasome can be assessed by treatment with proteasomal inhibitors like MG132 (purple line). Blocking both autophagic-lysosomal and proteasomal activity simultaneously will abrogate both main sources of LLPD, thus resulting in minimal loss of 14C-labelled intact protein (black line). Note that the rise and fall in the curves are schematic and purely intended for illustrative purposes–they are not intended to indicate exact details in the kinetics of long-lived protein labelling and/or degradation. See text for a more detailed description of each of the protocol steps and for representative examples of data.
Materials and Reagents
In the current protocol we use:
- Pipette tips (Thermo Fisher ART Barrier tips) (VWR, catalog numbers: 732-2223 (0.5-20 µl), 732-2207 (1-200 µl), and 732-2355 (100-1,000 µl))
- 75 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353136 )
- 24-well tissue culture plates (Corning, Falcon®, catalog number: 353047 )
- Microcentrifuge tubes (VWR, catalog number: 211-2130 )
- Scintillation vials (PerkinElmer, catalog number: 6000292 )
- Nitrile gloves (VWR, catalog number: 112-2372 )
- 50 ml tube (VWR, catalog number: 525-0402 )
- 0.45 µm filter (VWR, catalog number: 514-0075 )
- LNCaP cells (ATCC, catalog number: CRL-1740 )
- U2OS cells (ATCC, catalog number: HTB-96 )
- VCaP cells (ATCC, catalog number: CRL-2876 )
- Huh7 cells (Nakabayashi et al., 1982) (Kindly provided by Dr. Line M. Grønning-Wang, Oslo, Norway)
- PBS (Thermo Fisher Scientific, GibcoTM, catalog number: 20012019 )
- 0.25% Trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056 )
- RPMI 1640 (Thermo Fisher Scientific, GibcoTM, catalog number: 21875091 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F7524 )
- [1-14C] L-valine, 45 mCi/mmol, 0.1 mCi/ml (Vitrax, catalog number: VC 308 )
- Poly-D-lysine (Sigma-Aldrich, catalog number: P6407-10X5MG )
- For RNAi reverse transfection:
Lipofectamine RNAiMAX (Thermo Fisher Scientific, InvitrogenTM, catalog number: 13778150 )
Ambion SilencerTM select siRNAs (negative control ‘siCtrl’, Thermo Fisher Scientific, InvitrogenTM, catalog number: 4390843 ; siULK1, s15964; siULK2, s18706)
Opti-MEM reduced serum medium (Thermo Fisher Scientific, GibcoTM, catalog number: 11058021 ) - Earle’s balanced salt solution (EBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 24010043 )
- Scintillation liquid (PerkinElmer, catalog number: 6013199 )
- DMSO (Sigma-Aldrich, catalog number: D2650 )
- Bafilomycin A1 (Enzo Life Sciences, catalog number: BML-CM110-0100 )
- Ammonium chloride (NH4Cl) (Sigma-Aldrich, catalog number: A4514 )
- SAR-405 (Magento, ApexBio, catalog number: A8883 )
- Torin1 (Tocris Bioscience, catalog number: 4247 )
- Non-radioactive L-valine (Sigma-Aldrich, catalog number: V0513 )
- Bovine serum albumin (BSA) (VWR, catalog number: 422361V )
- Trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T0699 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 60377 )
- 200 mM cold L-valine (see Recipes)
- RPMI 1640/10%FBS (see Recipes)
- 1 mg/ml poly-D-lysine (see Recipes)
- 1 mM Torin1 (see Recipes)
- PBS/2%BSA (see Recipes)
- 25% TCA (see Recipes)
- 0.2 M KOH (see Recipes)
- 0.2 mM Bafilomycin A1 (see Recipes)
- 160 mM NH4Cl (see Recipes)
Equipment
- Pipettes (FinnpipetteTM F2 GLP Kit) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4701070 )
- Humidified incubator
- Tube rotator (VWR, catalog number: 444-0502 )
- Plate shaker (Grant Instruments, model: PMS-1000i )
- Magnetic stirrer (IKA, catalog number: 0003810001 )
- Autoflow IR Direct Heat CO2 incubator (NuAire, model: NU-5510E )
- Vortexer (Denville Scientific, catalog number: S7030 )
- Benchtop centrifuge with cooling (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75002430 )
- Scintillation counter (Liquid Scintillation Analyzer) (Packard, model: 1600 TR )
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Luhr, M., Sætre, F. and Engedal, N. (2018). The Long-lived Protein Degradation Assay: an Efficient Method for Quantitative Determination of the Autophagic Flux of Endogenous Proteins in Adherent Cell Lines. Bio-protocol 8(9): e2836. DOI: 10.21769/BioProtoc.2836.
分类
生物化学 > 蛋白质 > 降解
细胞生物学 > 基于细胞的分析方法 > 自噬活性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link