- EN - English
- CN - 中文
A Modified Low-quantity RNA-Seq Method for Microbial Community and Diversity Analysis Using Small Subunit Ribosomal RNA
改良的低量 RNA-Seq法通过检测小亚基核糖体RNA以分析微生物群落和多样性
发布: 2018年05月05日第8卷第9期 DOI: 10.21769/BioProtoc.2828 浏览次数: 6152
评审: Dennis NürnbergStefan de VriesShyam Solanki
Abstract
We propose a modified RNA-Seq method for small subunit ribosomal RNA (SSU rRNA)-based microbial community analysis that depends on the direct ligation of a 5’ adaptor to RNA before reverse-transcription. The method requires only a low-input quantity of RNA (10-100 ng) and does not require a DNA removal step. Using this method, we could obtain more 16S rRNA sequences of the same regions (variable regions V1-V2) without the interference of DNA in order to analyze OTU (operational taxonomic unit)-based microbial communities and diversity. The generated SSU rRNA sequences are also suitable for the coverage evaluation for bacterial universal primer 8F (Escherichia coli position 8 to 27), which is commonly used for bacterial 16S rRNA gene amplification. The modified RNA-Seq method will be useful to determine potentially active microbial community structures and diversity for various environmental samples, and will also be useful for identifying novel microbial taxa.
Keywords: RNA-Seq (RNA-Seq)Background
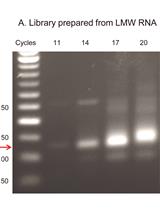
Ribosomal RNA (rRNA) accounts for more than 90% of the total microbial RNA, and is suitable for the analysis of microbial communities as an indicator of microbial physiological activity to synthesize proteins (Blazewicz et al., 2013). The study of microbial community transcripts, including rRNA and mRNA, in a particular environment (Double RNA metatranscriptomics) has advantages in providing both functional and taxonomic information on microbes (Urich et al., 2008), but has failed to perform OTU-based community comparisons. Although diversity indices could be calculated and compared using the V3 region of 16S rRNA sequences when gel-extracted SSU rRNA is analyzed, only a third of the resulting 16S rRNA sequences were found to be suitable for such analyses (Li et al., 2014). Besides, such method usually requires high quantities of RNA (Li et al., 2014). We recently developed a modified RNA-Seq method that uses an immediate adaptor ligation step at the 5’ end of the RNA prior to reverse transcription. Consequently, we can obtain more 16S RNA reads which can be used for OTU-based community and diversity analysis especially regarding low-RNA-yield samples such as tap water, shower curtain and human skin (Yan et al., 2017), and also mudflat sediment samples (Yan et al., 2018).
Materials and Reagents
- RNA extraction
- RNA-Seq library preparation
- PCR tubes (Corning, Axygen®, catalog number: PCR-02-L-C )
- RNA-Seq Library Preparation Kit for Whole Transcriptome Discovery–Illumina Compatible (Gnomegen, catalog number: K02421-T )
- DNase/RNase free water (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9932 )
- QubitTM RNA HS Assay Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32855 )
- RNaseOUTTM Recombinant Ribonuclease Inhibitor (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10777019 )
- QubitTM dsDNA HS Assay Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32854 )
- Gnome Size Selector (Gnomegen, catalog number: R02424S )
- PCR tubes (Corning, Axygen®, catalog number: PCR-02-L-C )
- Other materials
- Pipette tips (10 μl, 200 μl, 1,000 μl) (Corning, Axygen®, catalog numbers: TF-300-R-S , TF-200-R-S , TF-1000-R-S )
- 1.5 ml Eppendorf tubes (Eppendorf, catalog number: 022363204 )
- 100% ethanol
- RNase-free 70% ethanol
- Agarose (Biowest, catalog number: 111860 )
- 50x TAE Buffer (Sangon Biotech, catalog number: B548101-0500 )
- DL2,000 DNA Marker (Takara Bio, catalog number: 3427A )
- Pipette tips (10 μl, 200 μl, 1,000 μl) (Corning, Axygen®, catalog numbers: TF-300-R-S , TF-200-R-S , TF-1000-R-S )
Equipment
- Pipettes (0-10 μl, 10-100 μl, 100-1,000 μl) (Eppendorf, catalog numbers: k03030 , k03031 , k03032 )
- Vortex-genieTM 2 (QIAGEN, model: Vortex-Genie® 2 )
- Fresco 17 centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM FrescoTM 17 )
- Qubit 2.0 Fluorometer (Thermo Fisher Scientific, model: Qubit 2.0 )
- Automated thermal cycler TP600 (Takara Bio, model: TP600 )
- MagneSphere® Technology Magnetic Separation Stand (twelve-position) (Promega, catalog number: Z5342 )
- Electrophoresis apparatus (Bio-Rad Laboratories)
- Gel imaging system
Software
- Sickle v1.33 (Joshi and Fass, 2011)
- Mothur v1.33.3 (Schloss et al., 2009)
- QIIME v1.8.0 (Caporaso et al., 2010)
- MIPE (Zou et al., 2017)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Yan, Y., Zhu, T., Zou, B. and Quan, Z. (2018). A Modified Low-quantity RNA-Seq Method for Microbial Community and Diversity Analysis Using Small Subunit Ribosomal RNA. Bio-protocol 8(9): e2828. DOI: 10.21769/BioProtoc.2828.
分类
微生物学 > 群落分析 > RNA-Seq
分子生物学 > RNA > RNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link