- EN - English
- CN - 中文
Metal-tagging Transmission Electron Microscopy for Localisation of Tombusvirus Replication Compartments in Yeast
使用金属标签透射电子显微技术定位酵母菌中番茄丛矮病毒复制区
发布: 2018年04月20日第8卷第8期 DOI: 10.21769/BioProtoc.2822 浏览次数: 7417
评审: David PaulMirko CorteseAnonymous reviewer(s)

相关实验方案

诱导型HIV-1库削减检测(HIVRRA):用于评估外周血单个核细胞中HIV-1潜伏库清除策略毒性与效力的快速敏感方法
Jade Jansen [...] Neeltje A. Kootstra
2025年07月20日 2460 阅读
Abstract
Positive-stranded (+) RNA viruses are intracellular pathogens in humans, animals and plants. To build viral replicase complexes (VRCs) viruses manipulate lipid flows and reorganize subcellular membranes. Redesigned membranes concentrate viral and host factors and create an environment that facilitates the formation of VRCs within replication organelles. Therefore, efficient virus replication depends on the assembly of specialized membranes where viral macromolecular complexes are turned on and hold a variety of functions. Detailed characterization of viral replication platforms in cells requires sophisticated imaging approaches. Here we present a protocol to visualize the three-dimensional organization of the tombusvirus replicase complex in yeast with MEtal-Tagging Transmission Electron Microscopy (METTEM). This protocol allowed us to image the intracellular distribution of the viral replicase molecules in three-dimensions with METTEM and electron tomography. Our study showed how viral replicase molecules build replication complexes within specialized cell membranes.
Keywords: Metal-tagging transmission electron microscopy (金属标签透射电子显微镜技术)Background
Replication of positive-stranded RNA viruses depends on the remodeling of cellular membranes. Intracellular membranes serve as a structural scaffold for VRC assembly, provide essential lipids and co-factors that modulate the activity of the viral replicase and protect the viral RNA from the antiviral defenses of the host (Miller and Krijnse-Locker, 2008; den Boon et al., 2010; Nagy and Pogany, 2011; Nagy, 2016). The architecture of replication organelles with active VRCs has been observed by electron microscopy. VRCs assemble in single membrane vesicles or ‘spherules’, tubulovesicular cubic membranes, double membrane vesicles (DMV) or planar oligomeric arrays (de Castro et al., 2013). Spherules are often observed in cells infected by RNA viruses. They form by invagination in a variety of organelles and have a narrow opening to the cytosol (den Boon et al., 2010).
Tomato bushy stunt virus (TBSV) is a small (+) RNA virus that belongs to the Tombusviridae, a family of viruses that infect plants. TBSV has recently emerged as a model virus to study viral replication and virus-host interactions using the yeast Saccharomyces cerevisiae as a model host (Nagy and Pogany, 2011). Studies of S. cerevisiae infected with plant viruses have facilitated the identification of numerous factors needed for viral replication (Nagy, 2008). Tombusviruses encode five proteins including two replication proteins, p92pol and p33. p92pol is the RNA-dependent RNA polymerase. The auxiliary protein p33 is an RNA chaperone that facilitates the recruitment of the viral RNA to the site of replication, in the cytosolic face of peroxisome membranes (McCartney et al., 2005; Jonczyk et al., 2007).
Understanding the biogenesis and functional architecture of VRCs in cell membranes is challenging and it requires sophisticated imaging techniques. Transmission electron microscopy (TEM) has contributed to our understanding of the architecture and organization of macromolecular assemblies in cells. However, methods to unambiguously identify proteins within the environment of the cell are lagging behind. In our lab, we have developed a new labeling method named METTEM from MEtal-Tagging Transmission Electron Microscopy. This method uses the metal-binding protein metallothionein (MT) as a genetically clonable tag for electron microscopy (Diestra et al., 2009; Risco et al., 2012). Mouse MT 1 is a small, 61-amino acid protein with 20 cysteine residues that bind gold atoms very efficiently. MT fused to a protein of interest and treated with gold salts, builds an electron-dense gold nanocluster of around 1 nm diameter, easily visualized by electron microscopy (Mercogliano and DeRosier, 2006 and 2007) (Figure 1). METTEM allows identification and localization of intracellular proteins with high specificity and exceptional sensitivity at molecular-scale resolution ( Diestra et al., 2009; Delebecque et al., 2011; Bouchet-Marquis et al., 2012; Risco et al., 2012; Barajas et al., 2014a; de Castro Martin et al., 2017; Fernandez de Castro et al., 2017).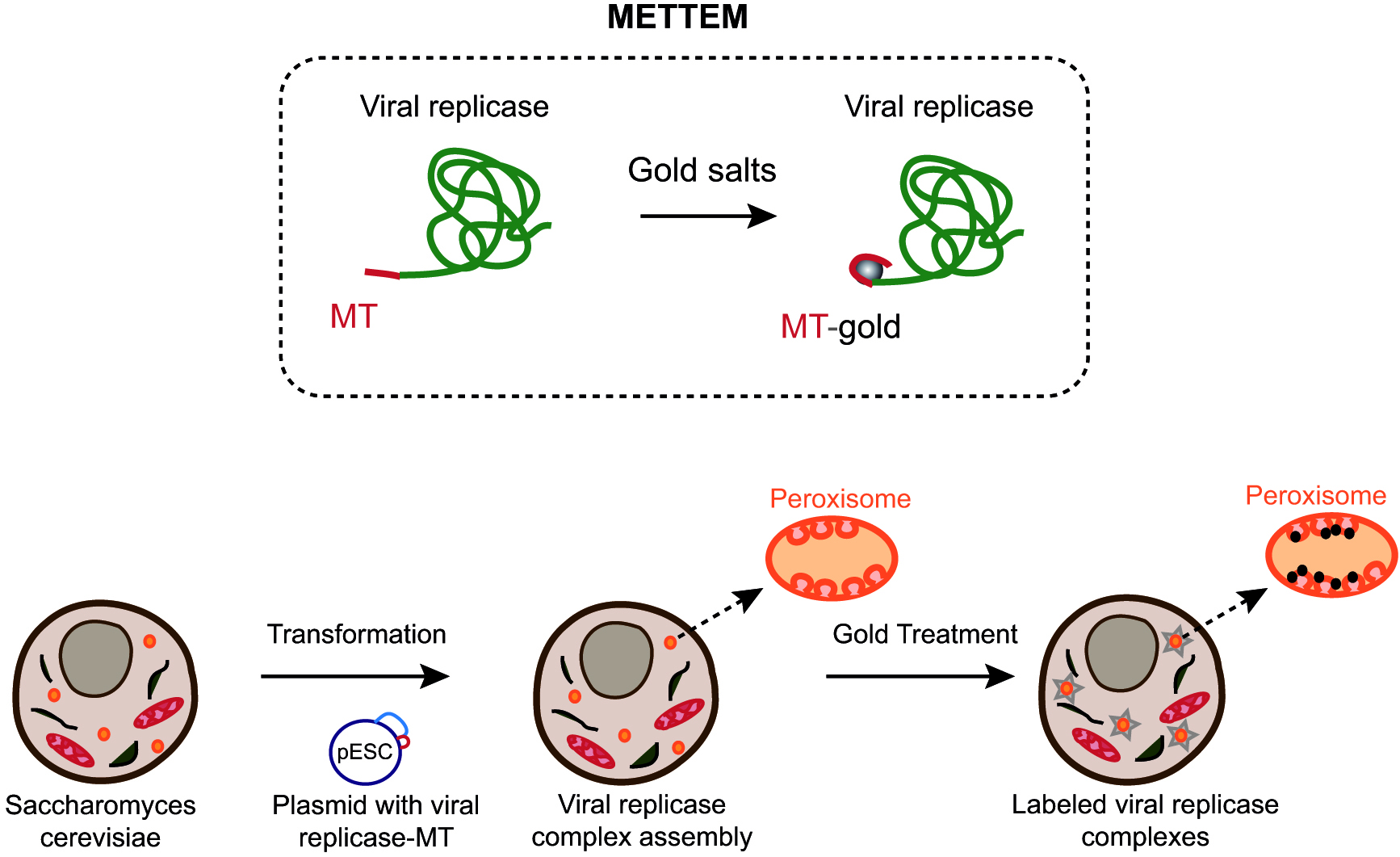
Figure 1. Imaging viral replicase complexes with METTEM. Viral replicase protein is fused with metallothionein (MT) and expressed in yeast cells (Fernandez de Castro et al., 2017). MT-tagged viral replicase molecules assemble VRCs in peroxisome membranes. Cells are incubated with gold salts in vivo and MT-gold-replicase molecules are visualized by electron microscopy.
Here we described a protocol to visualize TBSV replicase molecules in VRCs using METTEM (Figure 2). The combination of this technology with electron tomography allowed us to study the distribution of replicase molecules in the viral replication compartment in three-dimensions (3D). Due to the high sensitivity of the method we could distinguish different states of aggregation of the viral replicase molecules in situ. This methodology can be used to detect any protein of interest in different subcellular locations of bacteria, yeast and mammalian cells. Furthermore, one advantage of this electron microscopy approach is that it can be used to study many different viruses in a variety of cell types by visualizing the MT tag incorporated in either complete viral particles or their proteins. This method has revealed virus-induced structures not seen before, as reported for Rubella virus, Tombusvirus and influenza virus (Risco et al., 2012; de Castro Martin et al., 2017; Fernandez de Castro et al., 2014 and 2017). 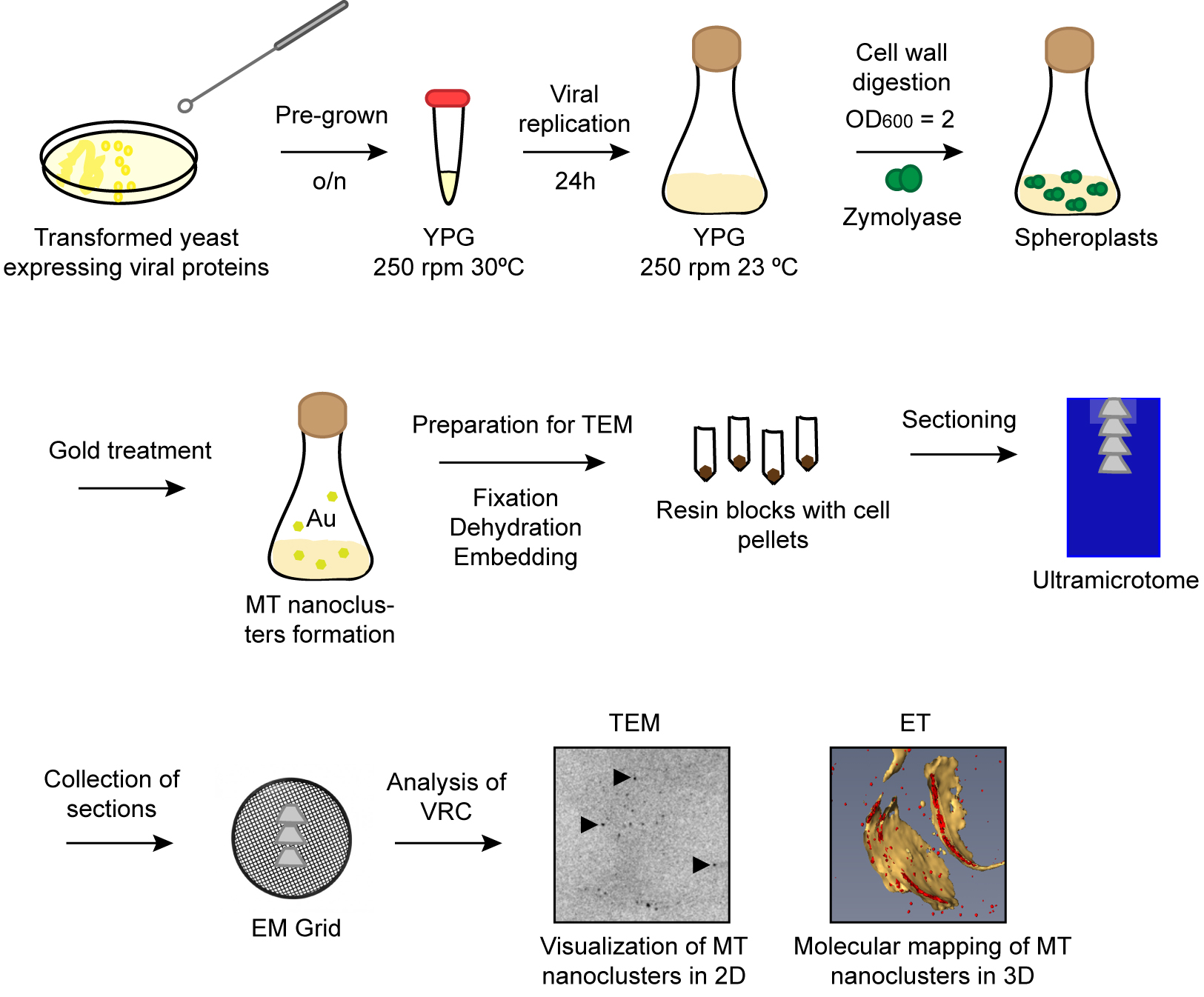
Figure 2. Schematic workflow of the protocol. Pre-grown transformed yeast cells are incubated overnight in YPG. Next day viral replication is induced during 24 h at 23 °C. Cells are treated with zymolyase to obtain spheroplasts. Spheroplasts are incubated with gold salts to build nanoclusters in MT tags. Cell pellets are dehydrated and embedded in resin. Serial sections are transferred to EM grids and imaged by TEM.
Materials and Reagents
- Pipette tips
- Perfect loop (Electron Microscopy Sciences, catalog number: 70945 )
- 50 ml disposable centrifuge tubes
- Eppendorf tubes
- Sterile transfer pipettes
- Serum Acrodisc® 37 mm syringe filter (Pall, catalog number: 4525 )
- Gelatin capsule size 1 6.5 mm diameter-0.50 ml (TAAB, catalog number: C089/1 )
- GEM® Single Edge Blades 3-Facet 0.009"/0.23 mm (AccuTec Blades, catalog number: 62-0179-0000 )
- Saccharomyces cerevisiae yeast strains RS453 (MATa ade2-1 his3, 15 leu2-3, 112 trp1-1 ura3 52) and pah1Δnem1Δ (SwissProt ID for Nem1 is P38757) (pah1Δ::TRP1nem1Δ::HIS3 derivative of RS453) (Choi et al., 2011; Barajas et al., 2014b)
- Zymolyase® 20T (Arthrobacter luteus) (Amsbio, catalog number: 120491-1 )
- Gold(III) chloride ≥ 99.99% (Sigma-Aldrich, catalog number: 379948 )
- Glutaraldehyde 50% (TAAB, catalog number: G015 )
- Ethanol Dry (Merck, catalog number: 1009901001 )
- LR-White Resin Medium Grade Acrylic resin (TAAB, catalog number: L012 )
- BactoTM yeast extract (BD, BactoTM, catalog number: 212750 )
- BactoTM peptone (BD, BactoTM, catalog number: 211677 )
- D-(+)-Glucose (Sigma-Aldrich, catalog number: G8270 )
- Agar (Sigma-Aldrich, catalog number: 05038 )
- D-(+)-Galactose (Sigma-Aldrich, catalog number: G0750 )
- Lithium acetate 99.95% (Sigma-Aldrich, catalog number: 517992 )
- ssDNA (Deoxyribonucleic acid sodium salt from salmon testes) (Sigma-Aldrich, catalog number: D1626 )
- PEG MW3350 (Polyethylene glycol) (Sigma-Aldrich, catalog number: P4338 )
- Trizma® hemisulfate (Sigma-Aldrich, catalog number: T8379 )
- 1,4-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: DTT-RO)
Manufacturer: Roche Diagnostics, catalog number: 10197777001 . - Yeast nitrogen base without amino acids (Sigma-Aldrich, catalog number: Y0626 )
- Yeast Synthetic Drop-out Medium Supplements without tryptophan (Sigma-Aldrich, catalog number: Y1876 )
- D-Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Tris (Base) (Norgen Biotek, catalog number: 28029 )
- Paraformaldehyde EM (TAAB, catalog number: P026 )
- NaOH
- 10x PBS
- 1,4-Piperazinediethanesulfonic acid, Piperazine-1,4-bis(2-ethanesulfonic acid), Piperazine-N,N’-bis(2-ethanesulfonic acid) PIPES (Sigma-Aldrich, catalog number: P6757 )
- 4-(2-Hydroxyethyl) piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid) (HEPES) (Sigma-Aldrich, catalog number: H3375 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- YPD (see Recipes)
- YPG (see Recipes)
- Transformation mix (see Recipes)
- TSD reduction buffer (see Recipes)
- Spheroplast medium A (see Recipes)
- 4% paraformaldehyde (PFA) solution (see Recipes)
- PHEM solution, pH 6.9 (see Recipes)
Equipment
- 500 ml flask
- Pipettes
- Diamond Knife 45° DIATOME (Fedelco)
- QUANTIFOIL® R 3.5/1 100 Holey Carbon Films Grids Au 300 mesh (Quantifoil)
- Oven Memmert UN 55 (Memmert, model: UN55 ) (Genesys) equipped with a shaker
- Spectrophotometer 722N (Terra Universal, Laboratory Equipment, model: 722N )
- Centrifuge 5810 R (Eppendorf, model: 5810 R )
- Centrifuge miniSpin plus (Eppendorf, model: MiniSpin® plus )
- Ultrasonic Cleaner 1510 (Branson, model: 1510 )
- pH meter Basic 20 (HACH LANGE SPAIN, Crison, model: Basic 20 )
- Fume Hood (Flow-Tronic)
- Ultramicrotome (Leica Microsystems, model: Leica EM UC6 )
- Jeol JEM 1011 electron microscope operating at 100 kV (JEOL, model: JEM-1011 )
- FEI Tecnai G2 F20 (200 kV) electron microscope (FEI)
- Tecnai Spirit Twin (120 kV) electron microscope (FEI)
Software
- IMOD software
- Amira software
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Fernandez de Castro, I. and Risco, C. (2018). Metal-tagging Transmission Electron Microscopy for Localisation of Tombusvirus Replication Compartments in Yeast. Bio-protocol 8(8): e2822. DOI: 10.21769/BioProtoc.2822.
分类
微生物学 > 微生物-宿主相互作用 > 病毒
细胞生物学 > 细胞成像 > 电子显微镜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link