- EN - English
- CN - 中文
Generation of Luciferase-expressing Tumor Cell Lines
表达萤光素酶的肿瘤细胞系的生成
发布: 2018年04月20日第8卷第8期 DOI: 10.21769/BioProtoc.2817 浏览次数: 20083
评审: Jia LiShalini Low-NamShweta Garg
Abstract
Murine tumor models have been critical to advances in our knowledge of tumor physiology and for the development of effective tumor therapies. Essential to these studies is the ability to both track tumor development and quantify tumor burden in vivo. For this purpose, the introduction of genes that confer tumors with bioluminescent properties has been a critical advance for oncologic studies in rodents. Methods of introducing bioluminescent genes, such as firefly luciferase, by viral transduction has allowed for the production of tumor cell lines that can be followed in vivo longitudinally over long periods of time. Here we describe methods for the production of stable luciferase expressing tumor cell lines by lentiviral transduction.
Keywords: Lentivirus (慢病毒)Background
Paramount to tracking cells in vivo is the ability to detect them externally by minimally invasive methods. Enzymatic bioluminescence using luciferase derived from the firefly (Photinus pyralis) is a widely used method for image-based cell tracking in vivo. Bioluminescence has been used for a variety of in vivo application including the noninvasive imaging of reporter gene expression (Herschman, 2004), studying circadian rhythms (Southern and Millar, 2005), imaging cerebral strokes (Vandeputte et al., 2014), and for tracking genetically engineered T cells (Costa et al., 2001; Cheadle et al., 2010). Perhaps the field where bioluminescent cell lines have been most applicable is oncology where they have been instrumental for the monitoring tumor growth (Jenkins et al., 2005; Brennan et al., 2016; Byrne et al., 2016) and tumor metastasis (Rosol et al., 2003; Simmons et al., 2015) in mouse models. While some subcutaneously implanted tumors can be detected by palpation and measured with calipers, these methods are not effective for monitoring metastases or tracking tumors that disseminate widely, such as hematological malignancies that commonly grow in the bone marrow, lymph nodes and spleen.
Firefly luciferase oxides luciferin in the presence of molecular oxygen, magnesium and adenosine triphosphate to produce yellow-green light at 560 nm (Wilson and Hastings, 1998; Fraga, 2008). Benefits of luciferase bioluminescence in cell tracking include penetration of tissue for non-invasive monitoring and the re-usability of the enzymatic marker. Another advantage of luciferases is that most cells are not luminescent such that high signal-to-noise ratios can be achieved. A limitation bioluminescence is photon attenuation caused by intervening tissues, such as skin, bone, or hair.
Firefly luciferase is a single polypeptide specified by the luc gene that can be readily cloned into vectors used in gene delivery. Transient expression by plasmid transfection or non-integrating virus transduction limits the time over which cell tracking can be performed. This is especially problematic for oncology studies that may last several months. The ability of retroviruses to integrate into the genome is a key attribute that favors their use in producing stable cell lines. However, some oncoretroviruses, such as the Moloney murine leukemia virus can be limited by transgene silencing over time (Jähner et al., 1982). Further, retroviral vectors require cell division for genomic integration and can be inefficient at transducing highly differentiated cells such as neurons, dendritic cells, or resting lymphocytes.
For the purpose of making luciferase expressing cell lines, lentiviral retroviral vectors derived from human immunodeficiency virus-1 (HIV-1) are highly effective. An advantage of lentiviral vectors over other retroviral vectors, is their ability to integrate into the genome of non-dividing cells. This property makes them suitable gene delivery vehicles for targeting highly differentiated cells, such as neurons, dendritic cells and lymphocytes (Naldini et al., 1996). Lentiviruses also deliver very stable genomic integration and long-term transgene expression, to the extent that they have been used to make transgenic mice following embryo transduction (Lois et al., 2002).
In order to track hematologic tumor cells in an in vivo murine leukemia model, we made the FULGW lentiviral vector that co-expresses firefly luciferase (Luc) and enhanced green fluorescent protein (EGFP) for the purpose of B-cell lymphoma (A20) cell line transduction and stable clone production. The FULGW vector is based on a self-inactivating vector previously described by Miyoshi et al. (1998) that had been engineered to express the GFP reporter gene behind the human ubiquitin-C promoter by Lois et al. (2002), making the FUGW vector. To make FULGW, we replaced the EGFP sequence of FUGW with a Luc-IRES-EGFP sequence from rKat.Luc2.IRES.EGFP, previously developed by Cheadle et al. (2010).
FULGW contains unique elements that enhance gene integration and expression (Figure 1A). It encodes the human immunodeficiency virus-1 (HIV-1) flap element, giving it karyotropic properties that permit efficient genomic integration in non-replicating cells (Zennou et al., 2000). It contains the wood-chuck hepatitis virus posttranscriptional regulatory element (WPRE) that increases gene expression by transcript stabilization (Zufferey et al., 1999). In addition, it includes a 3’ self-inactivating long terminal repeat (3’ si-LTR) that contributes to maintaining it as a replication deficient virus. The 3’ si-LTR was developed by the deletion of a 133 bp region in the U3 region (ΔU3) of the 3’ LTR that renders the 5’ LTR of the integrated provirus transcriptionally inactive (Miyoshi et al., 1998).
Virus production is performed by the co-transfection of HEK-293T cells with the lentiviral plasmid (FULGW) and the two packaging plasmids, pCMV-ΔR8.91 and pCMV-VSVG (Figure 1B). HEK-293T cells are a human embryonic kidney cell line that stably expresses the CMV large T antigen, which greatly increases gene expression by the CMV promoter, generating robust virus production. pCMVΔR8.91 is an HIV-1 Gag and Polymerase (Pol) expression plasmid that was modified from the dR8.9 vector by deletion of four accessory HIV-1 gents, Vif, Vpr, Vpu, and Nef (Zufferey et al., 1997). pCMV-VSVG expresses the pantropic envelop (Env) protein derived from the vesicular stomatitis virus glycoprotein (VSVG) (Stewart et al., 2003). [*Note: Both FULGW and pCMV-ΔR8.91 are large plasmids and best grown in chemically competent recA1-deficient E. coli with high transformation efficiency such as One Shot TOP10 E. coli (Invitrogen) grown at 30 °C for 24-28 h]. Using the FULGW lentiviral vector packaged with these helper plasmids, we have produced multiple types of tumor cell lines on various genetic backgrounds that stably express luciferase and GFP for use in oncologic studies (Table 1).
Figure 1. Production of FULGW lentivirus and transduction of tumor cell lines. A. Diagram of key regions of the FULGW vector including the Luc-IRES-EGFP transgene. Transgene expression is driven by the human ubiquitin-C promoter. CMV (cytomegalovirus promoter), U5 (LTR unique 5’ region), R (LTR repeat region), HIV-1 flap (human immunodeficiency virus-1 flap element), Luc (firefly luciferase), IRES (intra-ribosomal element sequence), EGFP (enhanced green fluorescent protein), WPRE (wood-chuck hepatitis virus posttranscriptional regulatory element), si-LTR (self-inactivating LTR). B. FULGW is packaged and pseudotyped by lipophilic co-transfecting with pCMV-ΔR8.91 and pCMV-VSVG. Virus rich culture supernatant (SN) is collected at 48 and 72 h and virus is concentrated by ultracentrifugation. The concentrated virus is used to transduce tumor cell lines by spin-transduction in the presence of polybrene. Illustrated schematics make use of Motifolio templates (www.motifolio.com/).
Table 1. Luciferase-GFP expressing tumor cell lines produced by FULGW transduction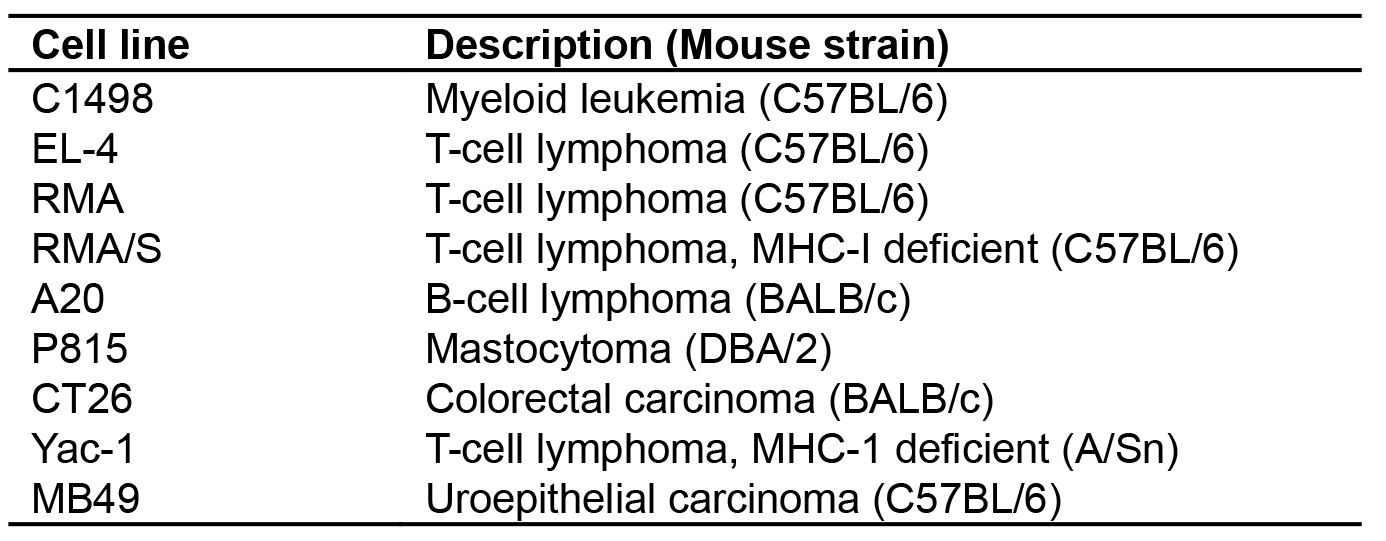
Materials and Reagents
- Pipette tips (USA Scientific, catalog numbers: 1110-3000 , 1110-1000 , 1111-2021 )
- T75 flask (Corning, Falcon®, catalog number: 353136 )
- 100 mm TC-treated Tissue Culture Dish (Corning, Falcon®, catalog number: 353003 )
- Sterile syringe 0.45 μm filter (VWR, catalog number: 28145-505 )
- Beckman ultra-clear 25 x 89 mm tubes (Beckman Coulter, catalog number: 344060 )
- Centricon Plus-70 unit (Merck, catalog number: UFC710008 )
- 15 ml Falcon tubes (Corning, Falcon®, catalog number: 352099 )
- 1.5 ml Eppendorf tubes (USA Scientific, catalog number: 1615-5500 )
- 12-well plates (Corning, catalog number: 3513 )
- 24-well plates (Corning, Costar®, catalog number: 3526 )
- 96-well plates (Greiner Bio One International, catalog number: 650185 )
- Sterile 500 ml 0.22 μm filter system (Corning, catalog number: 430758 )
- 29 ga. needles attached to 0.5 ml syringe (Terumo Medical Corporation, Elkton, MD, USA)
- BALB/c mice (THE JACKSON LABORATORY, catalog number: 000651 )
- HEK 293T (ATCC, catalog number: CRL-3216 )
- A20 B-cell lymphoma (ATCC, catalog number: TIB-208 )
- pCMV-VSVG plasmid (Addgene, catalog number: 8454 )
- pCMVΔR8.91 plasmid (Lifescience Market, catalog number: PVT2323 )
- pFULGW (Lentiviral luciferase-IRES-GFP plasmid, Available on request)
- Lipofectamine 2000 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668027 )
- Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 11058021 )
- Dulbecco’s phosphate buffered saline (DPBS) (Corning, catalog number: 21-031-CM )
- Trypsin EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056 )
- Polybrene (Sigma-Aldrich, catalog number: TR-1003-G )
- Propidium iodide (Sigma-Aldrich, catalog number: P4864-10ML )
- Bright-GloTM Luciferase Assay system (Promega, catalog number: E2610 )
- D-Luciferin, potassium salt (Gold Bio, catalog number: LUCK-1G )
- Isoflurane; Abbott Laboratories (Abbott Park, Illinois, USA)
- Fetal bovine serum (Corning, catalog number: 35-010-CV )
- DMEM–high glucose (Sigma-Aldrich, catalog number: D6429-500ML )
- L-Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Gelatin, 2% in H2O, tissue culture grade (Sigma-Aldrich, catalog number: G1393 )
- RPMI (Thermo Fisher Scientific, GibcoTM, catalog number: 11875093 )
- Penicillin/streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- D10 growth media (see Recipes)
- 2% gelatin (see Recipes)
- D-Luciferin stock solution (see Recipes)
Equipment
- Pipettes (Mettler-Toledo, Rainin, catalog numbers: 17008653 , 17008650 , 17008649 ; Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4641070N )
- Tissue culture hood
- Tissue culture incubator (Eppendorf, New Brunswick, model: Galaxy® 170 S )
- Fluorescent inverted microscope with GFP filter (Leica Microsystems, model: Leica DM IL LED )
- FACS-Canto flow cytometer (BD Biosciences)
- Microcentrifuge (Eppendorf, model: 5424 )
- Table-top centrifuge (Eppendorf, model: 5810 )
- Ultracentrifuge (Beckman Coulter, model: L8-80M ) equipped with an SW-28 rotor (Beckman Coulter, model: SW 28 )
- Balance (VWR, catalog number: 10204-990 )
- Xenogen IVIS Imaging System (Perkin Elmer, Hopkinton, MA, USA)
- Tabletop Laboratory Animal Anesthesia System (VetEquip, catalog number: 901806 )
- Autoclave
Software
- Acquisition software (CellQuest, BD Biosciences)
- Analysis software (FlowJo v9.3, TreeStar)
- Living Image Software (Caliper Life Sciences, Hopkinton, MA, USA)
- Graphing software (GraphPad Prism v7.0c, La Jolla, CA, USA)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Brennan, T. V., Lin, L., Huang, X. and Yang, Y. (2018). Generation of Luciferase-expressing Tumor Cell Lines. Bio-protocol 8(8): e2817. DOI: 10.21769/BioProtoc.2817.
分类
癌症生物学 > 通用技术 > 肿瘤形成 > 细胞分离和培养
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link