- EN - English
- CN - 中文
Microbial Mutagenicity Assay: Ames Test
微生物诱变分析:Ames试验
发布: 2018年03月20日第8卷第6期 DOI: 10.21769/BioProtoc.2763 浏览次数: 36116
评审: Modesto Redrejo-RodriguezAnonymous reviewer(s)
Abstract
The Microbial mutagenicity Ames test is a bacterial bioassay accomplished in vitro to evaluate the mutagenicity of various environmental carcinogens and toxins. While Ames test is used to identify the revert mutations which are present in strains, it can also be used to detect the mutagenicity of environmental samples such as drugs, dyes, reagents, cosmetics, waste water, pesticides and other substances which are easily solubilized in a liquid suspension. We present the protocol for conducting Ames test in the laboratory.
Keywords: Mutagenicity (诱变作用)Background
The Microbial Ames test is a simple, rapid and robust bacterial assay consisting of different strains and applications of Salmonella typhimurium/E. coli, used for ascertaining the mutagenic potential (Levin et al., 1982; Gupta et al., 2009). In 1975, Ames and his followers standardized the traditional Ames assay protocol and reappraised in 1980’s (Maron and Ames, 1983). Induction of new mutations replacing existing mutations allows restoring of gene function. The newly formed mutant cells are allowed to grow in the absence of histidine and form colonies, hence this test is also called as ‘Reversion assay’ (Ames, 1971). While traditional Ames test is quite laborious and time consuming for initial monitoring of mutagenic compounds, miniaturization of liquid suspension significantly impacted the usability by making it more convenient. The standard doses (2 µl, 5 µl, 10 µl, 50 µl and 100 µl) were set to evaluate the mutagenicity from lower to higher concentration (Hayes, 1982). Mice liver has been used as a tissue for preparing homogenate 9,000 x g (S9 hepatic fraction) whereas in S9 mix, hepatocytes are used to minimize the mammalian metabolic activation formed in the mice liver. In Ames bioassay, the sensitivity of a compound for mutagenicity is based on the knowledge that a substance which is mutagenic in the presence of liver enzymes metabolizing compound might be a carcinogen (Mathur et al., 2005).
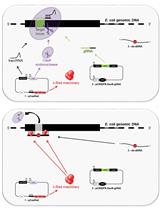
Genetic Approach: The Salmonella/E. coli tester strains: Several strains of Salmonella typhimurium have been used in Ames assay which requires histidine synthesis to assess the mutagenicity. In the histidine operon, each tester strain contains a different mutation. In addition to the histidine mutation, the standard tester strain of Salmonella typhimurium contains other mutations that greatly enhance their ability to detect the mutations (Figure 1). One of the mutations (rfa) causes partial loss of the lipopolysaccharides barrier that coats the surface of the bacteria and increases permeability to large molecules such as benzo[a]pyrene allowing not to penetrate in the normal cell wall (Mortelman and Zeiger, 2000). The mutagens present in the tested samples give rise to induced revertants on a minimal medium (absence of histidine). They are further used to observe revertants in previously mutated strains (that are not able to grow in a medium without histidine). The other mutation (uvrB) is a deletion mutation in which deletion of a gene, coding for the DNA excision repair system, causing gradually increased sensitivity in detecting many mutagens (Ames et al., 1973a). The reason behind this mutation is the deletion excising the uvrB gene emulsifying these bacteria requiring biotin for growth. The standard strains such as TA 97, TA 98, TA 100 and TA 102 contain the R-factor plasmid, pKM101. These R-factor strains are reverted by a number of mutagens that are detected weakly or not at all with the non R-factor parent strains (Ames et al., 1975a).
Figure 1. Genetic approach for assessing the mutagenicity in Salmonella strains (modified from https://en.wikipedia.org/wiki/Ames_test)
Many studies (Ames et al., 1975b; Levin et al., 1982) revealed that development of plasmid pKM101 in TA 1535 and TA 1538 strains leads to complement other isogenic strains such as TA 98, TA 100, TA 104 and TA 102. The his G46 mutation in TA 100 and TA 1535 codes for the first enzyme of histidine biosynthesis (hisG) (Ames et al., 1975b). This mutation, determined by DNA sequence analysis, substitutes proline (-GGG-) for leucine (-GAG-) in the wild type organism (Barnes et al., 1982). The tester strains TA 1535 and its R-factor derivative present in TA 100, detect mutagens which causes base-pair substitutions generally at one of these G-C pairs. The hisD3052 mutation in TA 1538 and TA 98 is in the hisD gene coding for histodinol dehydrogenase. TA 1538 and its R-factor derivative TA 98 detect various frameshift mutagens in repetitive sequences as ‘hot spots’ resulting in a frame shift mutation (Walker and Dobson, 1979; Shanabruch and Walker, 1980) (Table 1).
Table 1. Genotype of the Salmonella strain used for mutagenesis testing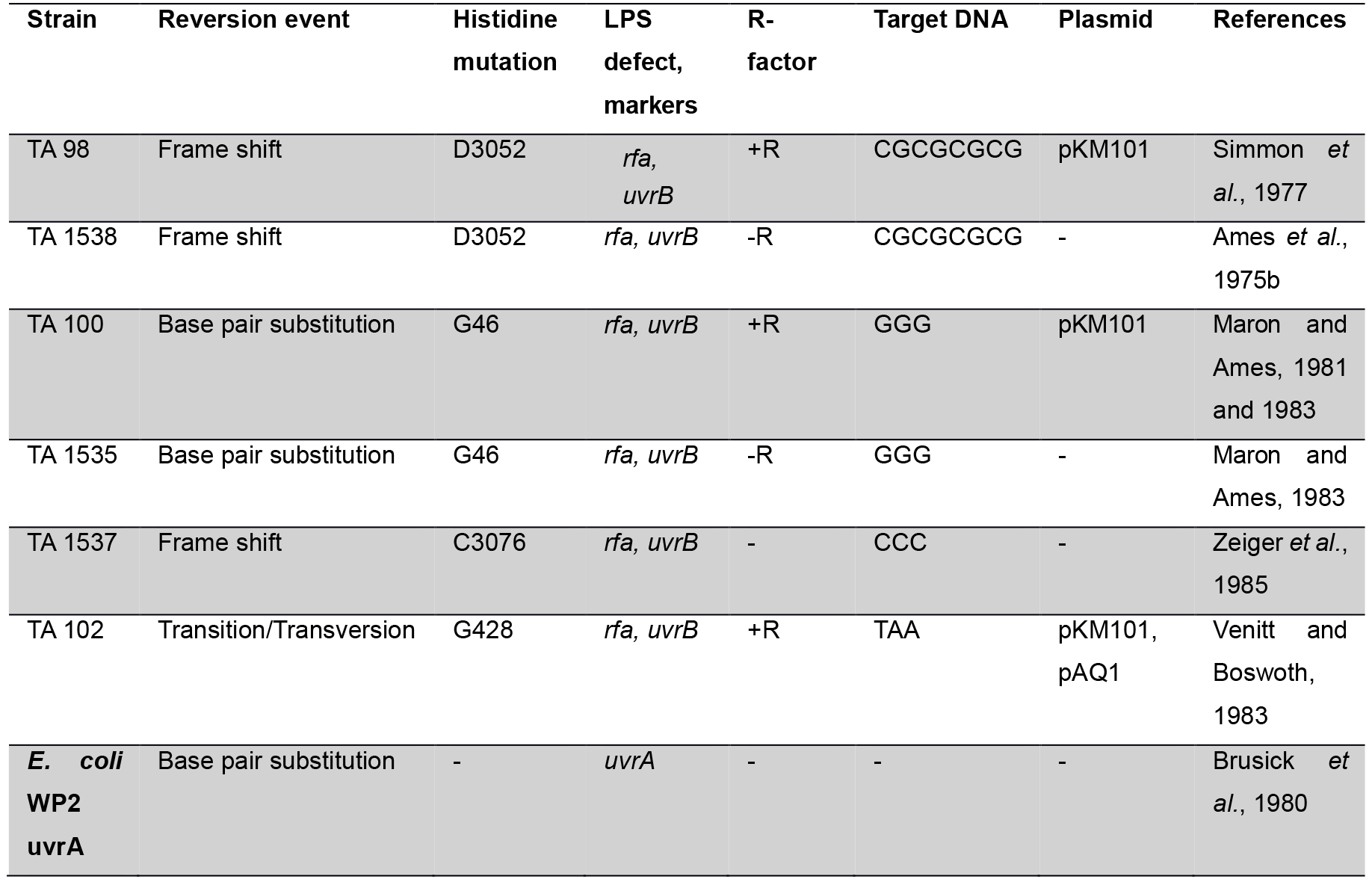
Levin et al. (1982) described a standard strain Salmonella typhimurium bacterium called TA 102 which was used to evaluate the effect of some compounds reacting with nucleotides AT. Tester strain TA102 containing nucleotides AT, present in hisG gene carrying plasmid pAQ1. There are certain mutagenic agents which are detected by TA 102 but not by TA 1535, TA 1537, TA 1538, TA 98 and TA 100 (Wilcox et al., 1990). Before performing experiment, a new set of fresh strains are prepared; and the genotypes are assessed (R-factor, His, rfa and uvrB mutations). For these, we refer readers to many excellent reviews (Walker, 1979; Czyz et al., 2002; Fluckiger-Isler et al., 2004).
Certain carcinogens present in active forms in biological reaction are easily catalyzed by cytochrome-P450. Metabolic activation system is absent in Salmonella, and in order to improve the potentiality of bacterial test systems, liver extracts of Swiss albino mice are used. This serves as a rich source in converting carcinogens to electrophilic chemicals that are incorporated to detect in vivo mutagens and carcinogens (Garner et al., 1972; Ames et al., 1973a). The crude liver homogenate as 9,000 x g S9 fraction contains free endoplasmic reticulum, microsomes, soluble enzymes and some cofactors set with S9 concentration to 10% (Franz and Malling, 1975). The oxygenase requires the reduced form of Nicotinamide Adenine Dinucleotide Phosphate (NADP) which is generally in situ by the action of glucose-6-phosphate dehydrogenase and reducing NADP both work as cofactors in assay (Prival et al., 1984; Henderson et al., 2000). While water is considered as a negative control, sodium azide, 2-nitrofluorine and mitomycin for TA 98, TA 100 and TA 102 without S9 metabolic activation and 2-anthramine with S9 hepatic fraction are used as positive controls for conducting the test (Table 2). Before performing the experiment, fresh solutions must be prepared.
Table 2. Positive controls with and without S9 metabolic activation (DeFlora et al., 1984)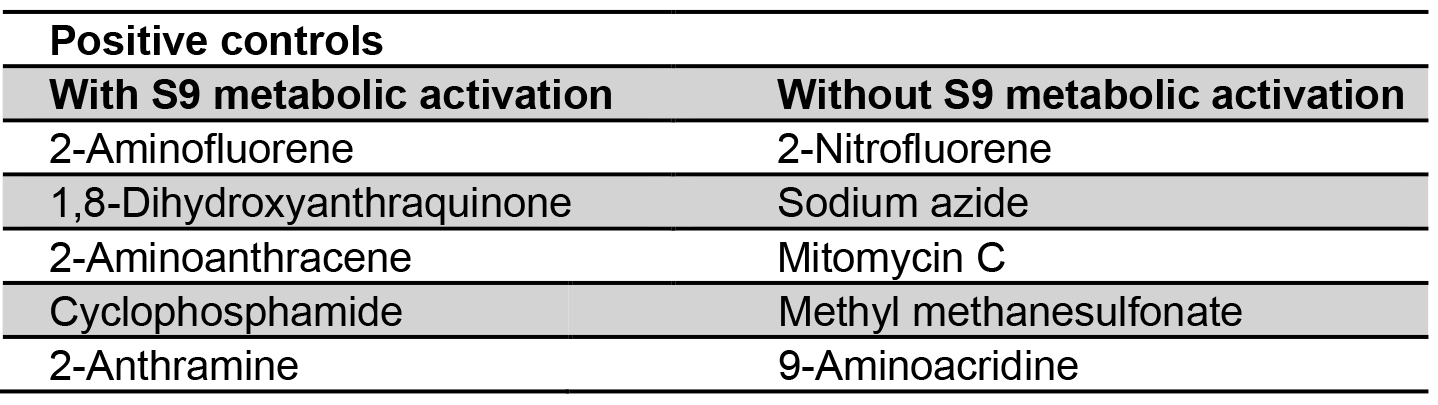
Spontaneous Reversion Control: Each strain of Salmonella contains a specific mutant range. Selection of solvents shows the effect on the frequency range of spontaneous mutant (Maron and Ames, 1983) (Table 3). The range of revertants varies in research laboratories. The spontaneous revertants are visible through unaided eyes (Figure 2).
Table 3. Spontaneous revertants control values for various strain types and number of revertants (Mortelmans and Stocker, 1979)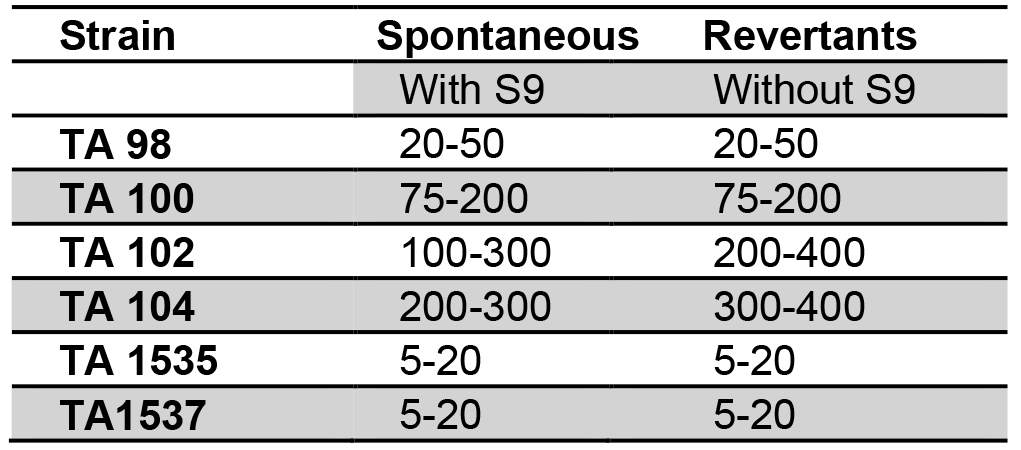
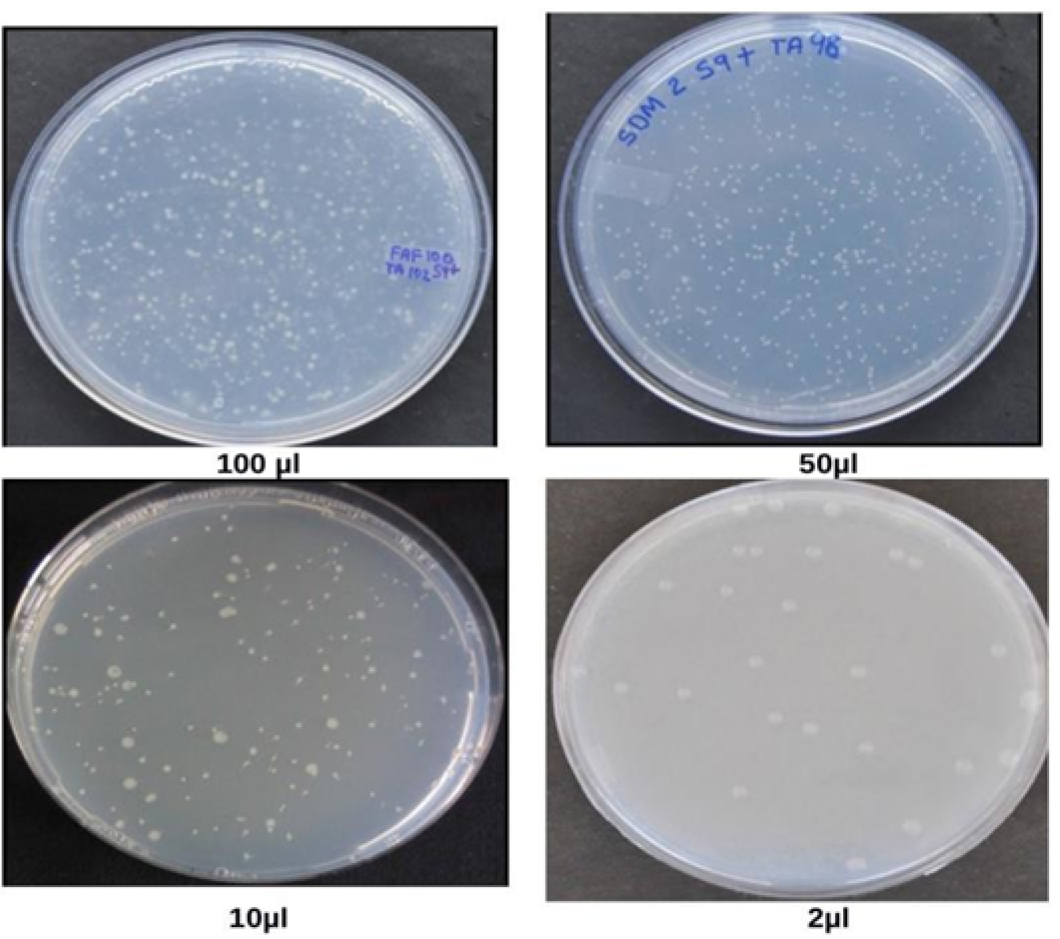
Figure 2. Spontaneous revertants colonies obtained after addition of waste water from health center in Salmonella mutagenicity assay at different concentrations, viz. 2 µl, 10 µl, 50 µl, 100 µl (Vijay, 2014)
Materials and Reagents
- Materials
- Tips (1,000 µl, 200 µl, 10 µl) (Tarsons)
- Sterile Petri plates (HiMedia Laboratories, catalog number: PW001 )
- Erlenmeyer flask and beaker (SchottDuran,10 ml, 250 ml, 500 ml)
- Eppendorf tubes (Tarsons,1.5 ml, 2.0 ml)
- Metal loop holder (metal loop Ch-2, HiMedia Laboratories, catalog number: LA012 )
- L shaped spreader(HiMedia Laboratories, catalog number: PW1085 )
- Tips (1,000 µl, 200 µl, 10 µl) (Tarsons)
- Mutagens
- Sodium azide (HiMedia Laboratories, catalog number: GRM1038 )
- 4-Nitroquinoline N-oxide (Sigma-Aldrich, catalog number: N8141 )
- 2-Aminofluorene (Sigma-Aldrich, catalog number: A55500 )
- Benzo(a)pyrene (Sigma-Aldrich, catalog number: B1760 )
- Mitomycin C (Roche Diagnostics, catalog number: 10107409001 )
- 2,4,7-Trinitro-9-fluorenone (Accustandard, catalog number: R-033S )
- 4-Nitro-o-phenylenediamine (Sigma-Aldrich, catalog number: 108898 )
- Sodium azide (HiMedia Laboratories, catalog number: GRM1038 )
- Reagents
- Oxoid nutrient broth No. 2 (Sigma-Aldrich, catalog number: 70123 )
Note: This product has been discontinued. - 70% ethanol
- Magnesium sulphate heptahydrate (MgSO4·7H2O) (HiMedia Laboratories, catalog number: RM683 )
- Citric acid monohydrate (HiMedia Laboratories, catalog number: GRM1008 )
- Potassium phosphate, dibasic (K2HPO4) (anhydrous) (Merck, catalog number: 61788005001730 )
- Sodium ammonium phosphate tetrahydrate (NaNH4HPO4·4H2O) (Sigma-Aldrich, catalog number: S9506 )
- D-biotin (HiMedia Laboratories, catalog number: TC096 )
- L-histidine (HiMedia Laboratories, catalog number: TC076 )
- Hydrochloric acid (HCI) (HiMedia Laboratories, catalog number: AS003 )
- Potassium chloride (KCl) (Merck, catalog number: 61753305001730 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (HiMedia Laboratories, catalog number: MB040 )
- Sodium dihydrogen phosphate monohydrate (NaH2PO4·H2O) (Merck, catalog number: 1063700050 )
- Disodium hydrogen phosphate (Na2HPO4) (HiMedia Laboratories, catalog number: TC051 )
- NADP (sodium salt) (HiMedia Laboratories, catalog number: RM392 )
- D-glucose-6-phosphate (monosodium salt) (Sigma-Aldrich, catalog number: G7879 )
- Ampicillin trihydrate (Sigma-Aldrich, catalog number: A6140 )
- Sodium hydroxide (NaOH) (Merck, catalog number: 106462 )
- Crystal violet (Sigma-Aldrich, catalog number: C6158 )
- Agar-Agar (Himedia Laboratories, catalog number: RM026 )
- Nutrient broth (HiMedia Laboratories, catalog number: M002 )
- Tetracycline (Sigma-Aldrich, catalog number: 87128 )
- Dimethylsulfoxide (HiMedia Laboratories, catalog number: TC185 )
- Vogel-Bonner medium E (50x) (see Recipes)
- 0.5 mM histidine/biotin solution (see Recipes)
- Salt solution (1.65 M KCl + 0.4 M MgCl2) (see Recipes)
- 0.2 M sodium phosphate buffer, pH 7.4 (see Recipes)
- 1 M Nicotinamide Adenine Dinucleotide Phosphate (NADP) solution (see Recipes)
- 1 M glucose-6-phosphate (see Recipes)
- Ampicillin solution (4 mg/ml) (see Recipes)
- Crystal violet solution (0.1%) (see Recipes)
- Minimal glucose plates (see Recipes)
- Histidine/Biotin plates (see Recipes)
- Ampicillin and tetracycline* plates (see Recipes)
- Nutrient agar plates (see Recipes)
- S9 mix (Rat Liver Microsomal Enzymes + Cofactors) (see Recipes)
- Sodium azide (see Recipes)
- Mitomycin (see Recipes)
- 2-Anthramine (see Recipes)
- Oxoid nutrient broth No. 2 (Sigma-Aldrich, catalog number: 70123 )
Equipment
- Orbital shaking incubator (Remi, model: RIS-24(BL) )
- Laminar Flow hood (Bio safety cabinet) (Deepak Meditech Pvt Ltd., Steri clean)
- Pipettes (Eppendorf, model: Research® plus, catalog number: 3120000062 , 1,000 μl; catalog number: 3120000046 , 200 μl; catalog number: 3120000020 , 10 μl)
- Vortex mixer (Labnet International, catalog number: S0100 )
- Hot water bath (Daiki Sciences, catalog number: KBLee2001 )
- Autoclave (TSC)
- Automatic Colony counter (Sonar)
- Refrigerator centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: Heraeus Biofuge Primo R )
- pH meter (Labindia Analytical Instruments, model: PICO pH Meter , catalog number: PC13330101)
- Tissue tearor (Bio Spec Products, catalog number: 985370-04 )
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Vijay, U., Gupta, S., Mathur, P., Suravajhala, P. and Bhatnagar, P. (2018). Microbial Mutagenicity Assay: Ames Test. Bio-protocol 8(6): e2763. DOI: 10.21769/BioProtoc.2763.
分类
微生物学 > 微生物遗传学 > 诱/突变
分子生物学 > DNA > DNA 损伤和修复
细胞生物学 > 细胞分离和培养 > 细胞生长
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link