- EN - English
- CN - 中文
Construction and Cloning of Minigenes for in vivo Analysis of Potential Splice Mutations
用于体内潜在的剪接突变分析的微小基因的构建及克隆
(*contributed equally to this work) 发布: 2018年03月05日第8卷第5期 DOI: 10.21769/BioProtoc.2760 浏览次数: 12134
评审: Nicoletta CordaniKate HannanPamela Maher

相关实验方案

一种改良的酰基-RAC方法分离视网膜棕榈酰蛋白质组,并通过LC-MS/MS进行后续检测
Sree I. Motipally [...] Saravanan Kolandaivelu
2023年04月20日 2638 阅读
Abstract
Disease-associated mutations influencing mRNA splicing are referred to as splice mutations. The majority of splice mutations are found on exon-intron boundaries defining canonical donor and acceptor splice sites. However, mutations in the coding region (exonic mutations) can also affect mRNA splicing. Exact knowledge of the disease mechanism of splice mutations is essential for developing optimal treatment strategies. Given the large number of disease-associated mutations thus far identified, there is an unmet need for methods to systematically analyze the effects of pathogenic mutations on mRNA splicing. As splicing can vary between cell types, splice mutations need to be tested under native conditions if possible. A commonly used tool for the analysis of mRNA splicing is the construction of minigenes carrying exonic and intronic sequences. Here, we describe a protocol for the design and cloning of minigenes into recombinant adeno-associated virus (rAAV) vectors for gene delivery and investigation of mRNA splicing in a native context. This protocol was developed for minigene-based analysis of mRNA splicing in retinal cells, however, in principle it is applicable to any cell type, which can be transduced with rAAV vectors.
Keywords: Minigene (微小基因)Background
A substantial portion of disease-associated mutations (at least 15%) are predicted to result in aberrant mRNA splicing (Cartegni et al., 2002; Singh and Cooper, 2012; Sterne-Weiler and Sanford, 2014). The ‘classical’ splice mutations are those affecting the canonical sequences defining the 5’- and 3’-splice sites (donor and acceptor splice sites, respectively). However, splice mutations can also occur in other non-coding and coding regions (Wang and Cooper, 2007; Scotti and Swanson, 2016). There is growing evidence that the frequency of splice mutations in coding regions (exonic mutations) has been underestimated (Julien et al., 2016; Soukarieh et al., 2016). Exonic splice mutations (i.e., point mutations, insertions or deletions) can induce exon skipping, intron retention or lead to the generation of novel donor or acceptor splice sites. Depending on the gene and exon composition, these mechanisms can have different impacts on the protein level ranging from haploinsufficiency to gain of function. Nevertheless, the exact knowledge of molecular mechanisms underpinning the disease-causing mutations is essential for developing optimal treatments.
mRNA splicing occurs in a highly cell type-specific manner, highlighting the need to analyze the impact of potential splice mutations in the tissue which is primarily affected by the mutation (Wang et al., 2008). Consequently, in the optimal case, mRNA splicing should be analyzed on the native gene and in the native tissue. This option, however, is rather demanding for several reasons:
1) It might require the elaborate generation of genetically modified human cell lines. This impedes a more systematic analysis of splice mutations for a single gene.
2) Many native cell types are highly specialized and their cultured counterparts (if available at all) do not reflect every morphological and molecular hallmark of the native cells including the composition and activity of the splicing machinery.
3) The generation of humanized animal models expressing the respective splice mutation in a given tissue is not only technically challenging, but also time-consuming and costly. Therefore, this approach also appears rather unsuitable for systematic testing of splice mutations for a given gene.
4) Often, native genes are too large to be cloned into classical expression vectors.
One alternative to circumvent a number of these obstacles is to use human minigenes designed for expression in appropriate animal models (e.g., mouse). We have evaluated this approach in recent studies addressing the effects of disease-associated mutations in different genes, e.g., PRPH2, on mRNA splicing (Becirovic et al., 2016b; Nguyen et al., 2016; Khan et al., 2017; Petersen-Jones et al., 2017). For stable and specific ectopic expression of the minigenes, we took advantage of rAAV vectors. These vectors are capable of transducing a variety of different cell types in vivo (Zincarelli et al., 2008; Lisowski et al., 2014). Furthermore, the design, cloning, production and purification of rAAV vectors can be completed in a few weeks and does not require elaborate technical equipment (Becirovic et al., 2016a).
Most native genes including PRPH2 exceed the limited packaging capacity of AAVs (approx. 4.7 kb) (Wu et al., 2010). Therefore, we designed PRPH2 minigenes lacking large intronic parts, which usually do not contain information required for correct mRNA splicing. For genes which do not contain large exon numbers or sizes, shortening of the intronic sections also allows for introducing the entire protein coding region into the rAAV vector-based minigenes.
This strategy (cf. Figure 1) was developed and evaluated to analyze the impact of known disease-linked mutations on mRNA splicing and protein expression in photoreceptor-specific genes, but should in principle also be transferable to other cell types.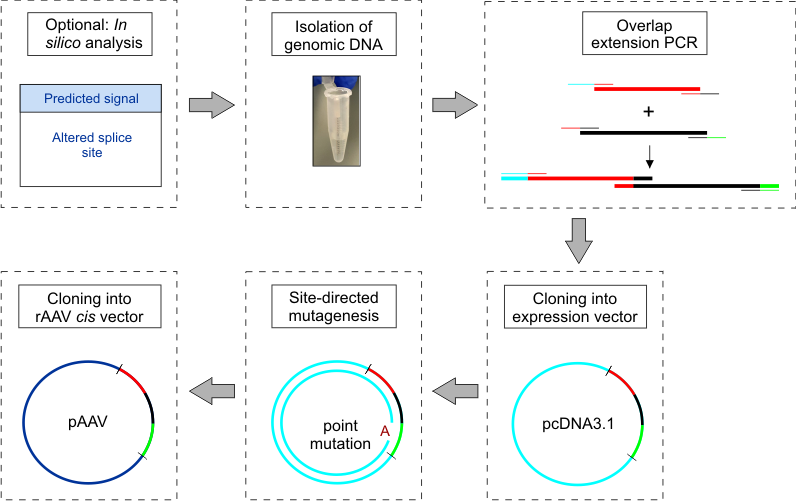
Figure 1. Workflow schematic for construction and cloning of minigenes
Materials and Reagents
- Isolation of human genomic DNA for cloning of minigenes
- 1.5 ml Eppendorf tubes (SARSTEDT, catalog number: 72.690.001 )
- Scalpel (Swann Morton, catalog number: 0510 )
- Gentra Puregene Buccal Cell Kit (QIAGEN, catalog number: 158845 )
- 2-Propanol (100%, ACS, ISO, Reag. Ph. Eur. grade)
- Ethanol (70%, ACS, ISO, Reag. Ph. Eur. grade)
- 1.5 ml Eppendorf tubes (SARSTEDT, catalog number: 72.690.001 )
- Overlap extension PCR for construction and cloning of minigenes
- PCR tubes (BRAND, catalog numbers: 781320 and 781334 )
- Expression vector of choice (e.g., pcDNA3.1 vector (+), Thermo Fisher Scientific, InvitrogenTM, catalog number: V79020 )
- Double distilled H2O (ddH2O)
- Herculase II Fusion DNA Polymerase (Agilent Technologies, catalog number: 600675 )
- dNTPs, 10 mM (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0192 )
- DNA Loading Dye (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0611 )
- GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM1331 )
- EDTA (VWR, catalog number: 20302.293 )
- Tris-(hydroxymethyl) aminomethane (VWR, catalog number: 103156X )
- Boric acid (VWR, catalog number: 20185.360 )
- QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704 )
- 1x TBE buffer (see Recipes)
- PCR tubes (BRAND, catalog numbers: 781320 and 781334 )
- Site-directed mutagenesis to introduce point mutations of interest into minigene
- PCR tubes (BRAND, catalog numbers: 781320 and 781334 )
- QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, catalog number: 210518 )
- Double distilled H2O (ddH2O)
- Peptone (Applichem, catalog number: 403898.1210 )
- Yeast extract (Applichem, catalog number: A1552 )
- NaCl (VWR, catalog number: 27810.364 )
- D-(+)-Glucose (Sigma-Aldrich, catalog number: 49159 )
- Agar (Sigma-Aldrich, catalog number: A5054 )
- Appropriate antibiotic for the plasmid vector (e.g., Ampicillin, Carl Roth, catalog number: K029.2 )
- LB agar plates containing the appropriate antibiotic for the plasmid vector (see Recipes)
- LB medium (see Recipes)
- PCR tubes (BRAND, catalog numbers: 781320 and 781334 )
- Subcloning of minigenes into rAAV vector
- Appropriate rAAV cis vector plasmid (e.g., pAAV-MCS2, Addgene, catalog number: 46954 )
- Appropriate rAAV cis vector plasmid (e.g., pAAV-MCS2, Addgene, catalog number: 46954 )
Equipment
- Pipettes (e.g., Eppendorf)
- Vortexer (Heidolph Instruments, model: Reax top )
- Incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: Heraeus B12 Function Line, catalog number: 50042307 )
- Microcentrifuge (Eppendorf, model: Centrifuge MiniSpin® , catalog number: 5452000018)
- Two water baths (Haake, catalog number: 003-2859 )
- Thermocycler (Thermo Fisher Scientific, model: ProFlexTM, catalog number: 4483636 )
- NanoDropTM (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: ND-2000C )
Software
- Human splicing finder software (e.g., http://www.umd.be/HSF3/)
- Tm Calculator Software (e.g., NEB Tm Calculator)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Riedmayr, L. M., Böhm, S., Michalakis, S. and Becirovic, E. (2018). Construction and Cloning of Minigenes for in vivo Analysis of Potential Splice Mutations. Bio-protocol 8(5): e2760. DOI: 10.21769/BioProtoc.2760.
分类
神经科学 > 感觉和运动系统 > 视网膜
发育生物学 > 形态建成 > 细胞结构
分子生物学 > RNA > RNA 剪接
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










