- EN - English
- CN - 中文
Visualization of RNA 3’ ends in Escherichia coli Using 3’ RACE Combined with Primer Extension
3'RACE联合引物延伸法检测大肠埃希氏菌RNA 3'末端
发布: 2018年03月05日第8卷第5期 DOI: 10.21769/BioProtoc.2752 浏览次数: 8983
评审: Gal HaimovichYi ZhangAnonymous reviewer(s)
Abstract
In this assay, 3’ RACE (Rapid Amplification of cDNA 3’ Ends) followed by PE (primer extension), abbreviated as 3’ RACE-PE is used to identify the mRNA 3’ ends. The following protocol describes the amplification of the mRNA 3’ ends at the galactose operon in E. coli and the corresponding visualization of the PCR products through PE. In PE, the definite primer is 5’ end-labeled using [γ-(32) P] ATP and T4 polynucleotide kinase, which anneals to the specific DNA molecules within the PCR product of the 3’ RACE. The conventional PE can only be used to locate the 5’ end of an mRNA transcript since reverse transcriptase (RTase) polymerizes only in the 5’ → 3’ direction. Thus, Taq polymerase is used instead of RTase, PCR is performed. Therefore, we are able to locate the 3’ end of the mRNA using this assay. The relative amount of the 3’ end can be directly visualized and quantified by way of separating DNA products in a denaturing 8% urea-PAGE (Polyacrylamide Gel Electrophoresis) gel. The exact position of the 3’ ends can be sequenced by comparison of these final DNA products with the corresponding DNA sequencing ladder.
Keywords: 3’ RACE (3' RACE)Background
The synthesis of the mRNA 3’ end is an important step in E. coli that produces a stable messenger RNA (mRNA). In eukaryotic cells, the mRNA 3’ end formation is through a cleavage from an internal phosphodiester bond, followed by the addition of a poly (A) tail; whereas in prokaryotic cells, the 3’ ends of mRNAs are generated by termination of transcription or by processing of the primary transcript (Altman and Robertson, 1973; Nudler and Gottesman, 2002; Zhao et al., 1999). Therefore, it is important to analyze the exact position and relative quantity of mRNA 3’ end to understand the mechanism of mRNA generation.
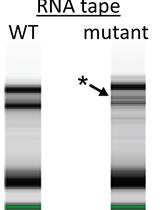
3’ RACE assay is a particular procedure to obtain the 3’ end sequence information of a defined RNA transcript (Sambrook and Russell, 2006). Generally, the experiment procedure starts with ligating the 3’ end of RNA to a synthetic RNA oligo, followed by the synthesis of cDNA using RTase and a complementary primer (3RP) to the RNA oligo. Subsequently, specific cDNA is amplified by PCR using the gene-specific primer and the primer, 3RP. Usually, RACE products are directly sequenced, however, based on our modified procedure, PCR products undergo another concluding step of primer extension (PE), which uses Taq polymerase instead of RTase. The labeled primer integrated into the PCR products are extended in a denaturing PAGE gel which makes us visualize and quantify each product. The scheme of 3’ RACE-PE is presented in Figure 1. The polycistronic gal operon encodes amphibolic enzymes for the amphibolism of the sugar D-galactose (Holden et al., 2003). Using this method, we have identified and quantitated the 3’ ends of the gal operon mRNAs in wild type and mutant strains (Lee et al., 2008; Wang et al., 2014 and 2015).
Figure 1. An illustration of the procedure 3’ RACE-PE in E. coli
Materials and Reagents
- Pipettes tips (DNase/RNase-free, Sorenson Bioscience)
- 1.5 ml centrifuge tube (SARSTEDT, catalog number: 72.690.001 )
- Cell culture flasks (Corning, catalog number: 3056 )
- 20 x 150 mm Test tube (Karter Scientific Labware Manufacturing, catalog number: 212W5 )
- Sephadex G-50 column (GE Healthcare, catalog number: 27-5330-01 )
- Whatman 3MM paper (GE Healthcare, catalog number: 3017-915 )
- Kodak CL-XPosure Film (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 34090 )
- Gloves (Ultra TEX Glove, Taeshin Bio Science, catalog number: UT-11 )
- E. coli strains (CGSC, the coli genetic stock center) (an example)
- Lysozyme (Roche Diagnostics, catalog number: 10837059001 )
- TRIZOL (Molecular Research Center, catalog number: TR 118 )
- Chloroform (Merck, Sigma-Aldrich, catalog number: C2432 )
- Isopropanol (Merck, Sigma-Aldrich, catalog number: I9516 )
- Ethanol (Merck, Sigma-Aldrich, catalog number: E7023 )
- RNA storage solution (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM7001 )
- Alkaline phosphatase (Takara Bio, catalog number: 2250A )
- DNase I (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2222 )
- RNasin® Ribonuclease Inhibitors (Promega, catalog number: N2111 )
- PCI (Phenol:Chloroform:Isoamyl Alcohol) Solution 25:24:1 (Merck, Sigma-Aldrich, catalog number: 77617 )
- Sodium acetate (Merck, Sigma-Aldrich, catalog number: S2889 )
- RNase and DNase-free water (Bioneer, catalog number: C-9011 )
- T4 RNA ligase (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2141 )
- The synthesized 3’ RACE RNA oligo: 3’-inverted deoxythymidine (3’-idT) RNA (Dharmacon)
- 3RP primer (5’AGCATGCGGCCGCTAAGAAC3’)
- dNTP mix (10 mM each) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0191 )
- HotStarTaq Plus DNA Polymerase (QIAGEN, catalog number: 203603 )
- Gal-specific primers (Table 1)
Table 1. Sequence of each galactose operon gene specific primers are listed
- T4 Polynucleotide Kinase (New England Biolabs, catalog number: M0201L )
- ATP, [γ-32P]- 6,000 Ci/mmol (PerkinElmer, catalog number: NEG002Z250UC )
- 5’ end labeled PE primer (5’ GAGAGTAGGGAACTGCCA 3’)
- 5x Developer (Vivid X-RAY DEVELOPER, Duksan (DS) Lab, catalog number: 0514_00004 )
- 5x Fixer (Vivid X-RAY RAPID FIXER, Duksan (DS) Lab, catalog number: 0514_00003 )
- Tryptone (BD, DifcoTM, catalog number: 211705 )
- Yeast extract (BD, DifcoTM, catalog number: 212750 )
- Sodium chloride (NaCl) (Merck, Sigma-Aldrich, catalog number: 746398 )
- Galactose (Merck, Sigma-Aldrich, catalog number: G5388 )
- Acrylamide (Merck, Sigma-Aldrich, catalog number: V900845 )
- Bis-acrylamide (Merck, Sigma-Aldrich, catalog number: V900301 )
- 5x TBE (Bioneer, catalog number: C-9002 )
- Urea (Merck, Sigma-Aldrich, catalog number: U5128 )
- Ammonium persulfate (Merck, Sigma-Aldrich, catalog number: 248614 )
- TEMED (Merck, Sigma-Aldrich, catalog number: T9281 )
- Formamide (Merck, Sigma-Aldrich, catalog number: F9037 )
- 0.5 M EDTA, pH 8.0 (Bioneer, catalog number: C-9007 )
- Xylene cyanol (Sigma-Aldrich, catalog number: X4126 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- 1 M Tris-HCl, pH 8.0 (Bioneer, catalog number: C-9006 )
- Sucrose (Merck, Sigma-Aldrich, catalog number: S7903 )
- Omniscript RT Kit (QIAGEN, catalog number: 205111 )
- Sigmacote (Sigma-Aldrich, catalog number: SL2 )
- LB + 0.5% galactose (see Recipes)
- 8% sequencing solution (see Recipes)
- 8% Urea-PAGE gel (see Recipes)
- 2x gel loading mix (see Recipes)
- Protoplasting buffer (see Recipes)
Equipment
- Automatic pipette aid (Thermo Fisher Scientific, Thermo ScientificTM, model: S1 Pipette Filler )
- Pipettes (Gilson Company, USA)
- 37 °C shaking incubator (Hanbaek Scientific Co., Korea (S))
- 37 °C and 42 °C heat block (FINEPCR, Korea (S))
- Spectrophotometer (BECKMAN COULTER, USA)
- Vortex-Genie 2 (Scientific Industries, model: Vortex-Genie 2 )
- 4 °C micro-centrifuge (Beckman Coulter, USA)
- NanoDrop 1000 (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 1000 )
- PCR machine (Bio-Rad Laboratories, USA)
- Shallow fixer tray (Bio-Rad Laboratories, USA)
- Sequencing gel electrophoresis apparatus (LABREPCO, USA)
- X-ray cassette (Duksan (DS) Lab, Korea (S))
- Power supply (Bio-Rad Laboratories, USA)
- Autoclave (Dong Won Scientific Corp, Korea (S))
Software
- ImageJ
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Wang, X., Jeon, H. J., Abishek N, M. P., He, J. and Lim, H. M. (2018). Visualization of RNA 3’ ends in Escherichia coli Using 3’ RACE Combined with Primer Extension. Bio-protocol 8(5): e2752. DOI: 10.21769/BioProtoc.2752.
分类
微生物学 > 微生物生物化学 > RNA
分子生物学 > RNA > 3' 末端分析
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link