- EN - English
- CN - 中文
Measurement of Arabidopsis thaliana Plant Traits Using the PHENOPSIS Phenotyping Platform
使用PHENOPSIS表型分析平台分析拟南芥植株性状
发布: 2018年02月20日第8卷第4期 DOI: 10.21769/BioProtoc.2739 浏览次数: 10860
评审: Tie LiuPaweł KrajewskiJason Liang Pin Ng
Abstract
High-throughput phenotyping of plant traits is a powerful tool to further our understanding of plant growth and its underlying physiological, molecular, and genetic determinisms. This protocol describes the methodology of a standard phenotyping experiment in PHENOPSIS automated platform, which was engineered in INRA-LEPSE (https://www6.montpellier.inra.fr/lepse) and custom-made by Optimalog company. The seminal method was published by Granier et al. (2006). The platform is used to explore and test various ecophysiological hypotheses (Tisné et al., 2010; Baerenfaller et al., 2012; Vile et al., 2012; Bac-Molenaar et al., 2015; Rymaszewski et al., 2017). Here, the focus concerns the preparation and management of experiments, as well as measurements of growth-related traits (e.g., projected rosette area, total leaf area and growth rate), water status-related traits (e.g., leaf dry matter content and relative water content), and plant architecture-related traits (e.g., stomatal density and index and lamina/petiole ratio). Briefly, a completely randomized (block) design is set up in the growth chamber. Next, the substrate is prepared, its initial water content is measured and pots are filled. Seeds are sown onto the soil surface and germinated prior to the experiment. After germination, soil watering and image (visible, infra-red, fluorescence) acquisition are planned by the user and performed by the automaton. Destructive measurements may be performed during the experiment. Data extraction from images and estimation of growth-related trait values involves semi-automated procedures and statistical processing.
Keywords: Phenotyping (表型)Background
Phenotyping of plant traits is an important aspect of plant sciences. It can be defined as a set of methodologies used to measure plant traits with certain accuracy and precision at different scales of organization (Fiorani and Schurr, 2013). A renaissance of plant phenotyping was brought by the development of automated phenotyping platforms and the creation of new tools for image analysis (Granier and Vile, 2014). Automated plant phenotyping is useful in many aspects of plant biology, such as ecophysiology (Vile et al., 2012), genetics (Bac-Molenaar et al., 2016), and molecular biology (Baerenfaller et al., 2012). It is then important for the broader readership to understand how these platforms work and what can be achieved. This protocol focuses on one particular installation, namely PHENOPSIS (Granier et al., 2006). PHENOPSIS is a custom-made phenotyping platform (growth chamber, automaton, and computer software), manufactured by Optimalog company and it is especially well-suited for analyses of small plants, such as Arabidopsis thaliana. The platform enables growing up to 504 A. thaliana plants simultaneously. Each plant is grown in a separate pot. Each pot can be automatically weighed and watered to a target value, thus it is feasible to monitor soil water content (SWC) individually and automatically adjust it. This feature of the platform makes it perfect for the application of water deficit treatments. PHENOPSIS automatically takes images of plant rosettes. Images can be of four types: RGB zenithal, RGB lateral, infrared, and fluorescence. In addition, many other non-destructive or destructive measurements are also available with some human involvement, e.g., transpiration, stomatal conductance, phenological stage, individual leaf area, epidermal cell density, extent of endoreduplication, and root weight. Because each of these measurements would require a separate lengthy manual, this protocol focuses on the basic platform operation, as well as measurements of leaf morphology and water status. The video of the platform in action is available online (http://bioweb.supagro.inra.fr/phenopsis/InfoBDD.php).
Materials and Reagents
- Double-sided tape (Scotch 12 x 6.3 mm dispensed, permanent, clear)
- Single-sided tape (Crystal clear tape Scotch 19 x 33 mm)
- Template sheets for leaf scanning
- Outer pots (APTE Society, http://www.apte.fr/)
- Inner perforated pots (outer pots hand perforated with a Dremel 4000)
- Pot labels (Point label, soft plastic 1.3 x 6 cm, BIER: http://www.fournitures-horticoles.com/magasin/catproduits.php?idgdf=9)
- Wooden toothpicks
- Microscope slide (Knittel Glass 76 x 26 mm, Starfrost)
- Pencil
- Fine permanent marker (Staedtler permanent, Lumacolor)
- Cylindrical containers for soil drying (50 x 50 mm, Servilab, catalog number: 8668770 )
- Paper bags for plant tissue drying (7 x 12 cm: http://www.beaumont-group.fr/produit/kraft-blanchi-frictionne-neutre-2/)
- Scalpel (Swann-Morton, Carbon Steel Surgical blades)
- Pincers (S MurrayTM Stainless Steel Watchmaker Forceps 11 cm, Fisher Scientific)
- A. thaliana seeds
- Agricultural soil (from Mauguio city, near Montpellier, France: N 43 37′ 01″, E 4 00′ 33″)
- Compost (Neuhaus N2)
- Clear nail polish (Maybelline New York, express Finish 40)
- Ammonium dihydrogen phosphate (H2PO4NH4) (VWR, catalog number: 21305.290 )
- Potassium nitrate (KNO3) (VWR, catalog number: 26869.291 )
- Fe (E.D.D.H.A) (SEQUESTRENE 138 FE 100 SG)
- Nitric acid (HNO3) 52% (VWR, catalog number: 20412.362 )
- Boric acid (H3BO3) (Merck, catalog number: 1.00165.0100 )
- Manganese(II) sulfate monohydrate (MnSO4·H2O) (VWR, catalog number: 25303.233 )
- Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, catalog number: C2857-250G )
Note: This product has been discontinued. - Zinc sulfate heptahydrate (ZnSO4·7H2O) (Avantor Performance Materials, J.T. Baker, catalog number: 4382-01 )
- Ammonium molybdate tetrahydrate, (NH4)6Mo7O24∙4H2O (Sigma-Aldrich, catalog number: A7302-100G )
- Nutrient solution (see Recipes)
- Microelement solution (see Recipes)
Equipment
- PHENOPSIS phenotyping platform (Figure 1)
- Growth chamber
- Robotic arm (custom made by Optimalog: https://www.optimalog.com/phenopsis)
- Precision balance (Precisa, model: XB620C )
- Watering tubes
- RBG camera (Prosilica GC1600 (Allied Vision Technologies, model: PROSILICA GC 1600 ) with Computar Varifocal Megapixel M3Z1228C-MP (CBC, Computar, model: M3Z1228C-MP ), 12-36 mm, monture C zoom lens)
- Controlling computer (Dell)
- Growth chamber
- Laptop computer for phenological stage notations (HP ProBook 650 G1 (HP Development, model: HP ProBook 650 G1 ), Intel Core i3-4000M Dual Core, 8 GB 1600 DDR3L 2DM)
- Desktop scanner (Epson, model: Epson Perfection 4990 Photo )
- Desktop computer for image analyses (HP Compaq Pro 6300 MT PC+ (HP Development, model: HP Compaq Pro 6300 Microtower ) Intel Core i5-3470 3.2 G 6 M HD 2500 CPU, GB DDR3-1600 DIMM (2 x 4 GB) RAM + 250 GB 7200 RPM 3.5 HDD, AMD Radeon HD 7450 DP)
- Tablet monitor with a pen (Wacom, model: Cintiq 22HD )
- Stereo microscope with camera attachment (Leitz DMRB, Manta G-201B camera)
- Green LED lamp (Bulb LED GU10 Spot 1 W = 10 W green)
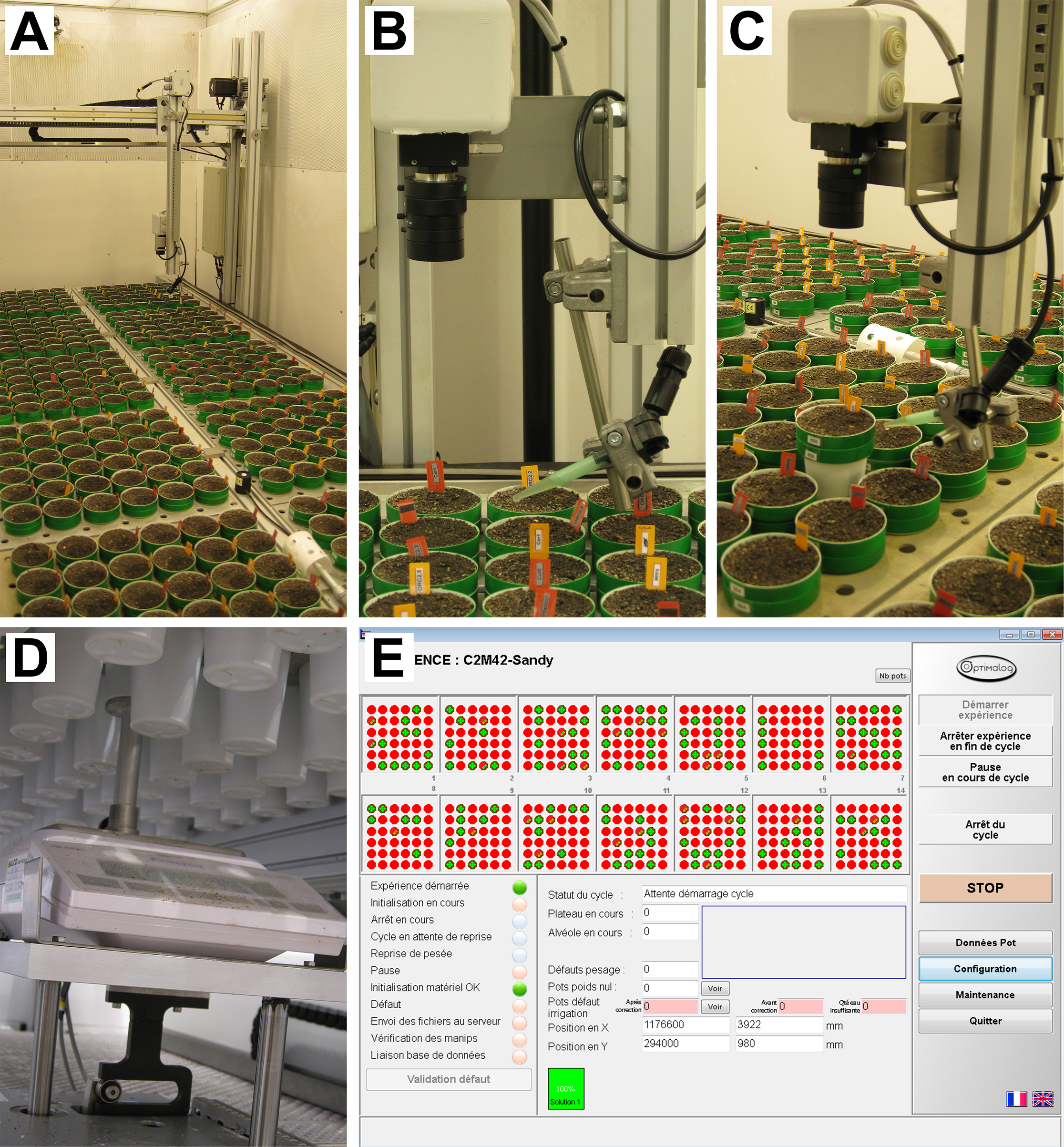
Figure 1. PHENOPSIS phenotyping platform. A. Overview of the growth chamber; B-C. Robotic arm with RBG camera and watering tube visible; D. Precision balance; E. A screenshot from Optimalog software controlling the platform.
Software
- Microsoft Excel
- ImageJ v1.51 (https://imagej.nih.gov/ij/) (Schneider et al., 2012)
- PHENOPSIS ImageJ macros (http://bioweb.supagro.inra.fr/phenopsis/MacroImageJ.php)
- Application for automaton control:
OPTIMA PLC software (https://optimalog.com/index.php?q=optimaplc_presentation) - Camera control: ProsilicaGigE (RGB Camera, Allied Vision); optionally: ThermaCAM Researcher (IR Camera, Flir), ImagingWin (Fluorescence camera, Walz)
Note: All the software is adapted by Optimalog to be used with Phenopsis. - R v3.4.1 (https://www.r-project.org/)
- Software for monitoring of environmental conditions:
Campbell Logger Net (https://www.campbellsci.com/loggernet)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Rymaszewski, W., Dauzat, M., Bédiée, A., Rolland, G., Luchaire, N., Granier, C., Hennig, J. and Vile, D. (2018). Measurement of Arabidopsis thaliana Plant Traits Using the PHENOPSIS Phenotyping Platform. Bio-protocol 8(4): e2739. DOI: 10.21769/BioProtoc.2739.
- Rymaszewski, W., Vile, D., Bediee, A., Dauzat, M., Luchaire, N., Kamrowska, D., Granier, C. and Hennig, J. (2017). Stress-related gene expression reflects morphophysiological responses to water deficit. Plant Physiol 174(3): 1913-1930.
分类
植物科学 > 植物生理学 > 植物生长
植物科学 > 植物生理学 > 非生物胁迫
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link