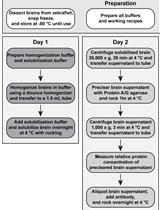
Fish Bile Clean-up for Subsequent Zymography and Mass Spectrometry Proteomic Analyses
鱼胆汁清除后进行酶谱和质谱蛋白质组学分析
发布: 2018年01月20日第8卷第2期 DOI: 10.21769/BioProtoc.2706 浏览次数: 7957
评审: Samik BhattacharyaPrashanth N SuravajhalaSalome Calado Botelho
引用
收藏
Cited by