- EN - English
- CN - 中文
The RiboPuromycylation Method (RPM): an Immunofluorescence Technique to Map Translation Sites at the Sub-cellular Level
RiboPuromycylation方法(RPM):一种在亚细胞水平绘制翻译位点图谱的免疫荧光技术
发布: 2018年01月05日第8卷第1期 DOI: 10.21769/BioProtoc.2669 浏览次数: 13578
评审: Alessandro DidonnaJosé M. DiasSalome Calado Botelho
Abstract
While isotopic labeling of amino acids remains the reference method in the field for quantifying translation rate, it does not provide any information on spatial localization of translation sites. The rationale behind developing the ribopuromycylation method (RPM) was primarily to map translation sites at the sub-cellular level while avoiding detection of newly synthesized proteins released from ribosomes. RPM visualizes actively translating ribosomes in cells via standard immunofluorescence microscopy in fixed and permeabilized cells using a puromycin-specific monoclonal antibody to detect puromycylated nascent chains trapped on ribosomes treated with a chain elongation inhibitor.
Keywords: Translation site (翻译位点)Background
For decades, isotopic labeling of amino acids has been considered as the gold standard for studying protein translation. Though this method has proven to be remarkably accurate for evaluating translation rates, it provides no information on the location of translating ribosomes. More recently, amino acids analogs have enabled fluorescent detection nascent chains (Dieterich et al., 2007). Nevertheless, nearly all of the detected signal comes from polypeptides released from ribosome. Our initial idea was to develop a method to label nascent chains while still tethered to translating ribosomes.
Puromycin (PMY) is an aminoglycoside antibiotic that mimics charged tRNATyr and incorporates into the ribosome A site. Consequently, PMY triggers premature translation termination by ribosome catalyzed-covalent incorporation into the nascent chain COOH-terminus (Pestka, 1971) followed by release of PMY-peptide. Polyclonal antibodies (Abs) to PMY were initially generated (Eggers et al., 1997) to detect puromycylated nascent chains released from ribosomes by immunoblotting and immunoprecipitation. Subsequently, fluorescent PMY was used to label nascent chains by microscopy (Starck et al., 2004). Schmidt et al. (2009) found that cells exposed to PMY generate a sufficient amount of PMY-terminated cell surface proteins to enable detection by live cell flow cytometry using a monoclonal Ab (mAb), providing a measure of translation rates. Importantly, none of these methods discriminate between ribosome-attached or released PMY-peptides and are all hindered to some extent as measures of translation by degradation of released proteins.
Having found that chain elongation inhibitors, such as cycloheximide (CHX) or emetine, prevent the release of PMY-nascent chain from ribosomes, we developed the ribopuromycylation method (RPM) that enables immunofluorescent detection of translation sites at the sub-cellular level (Figure 1) (David et al., 2011 and 2012b). This method has been used by our labs and many other labs to study translation in neurons (Biever et al., 2015; Perry et al., 2016; Williams et al., 2016), migrating cells (Willett et al., 2011), immune cells (Seedhom et al., 2016) and stressed or infected cells (Kedersha et al., 2016; Emmott et al., 2017; Roth et al., 2017).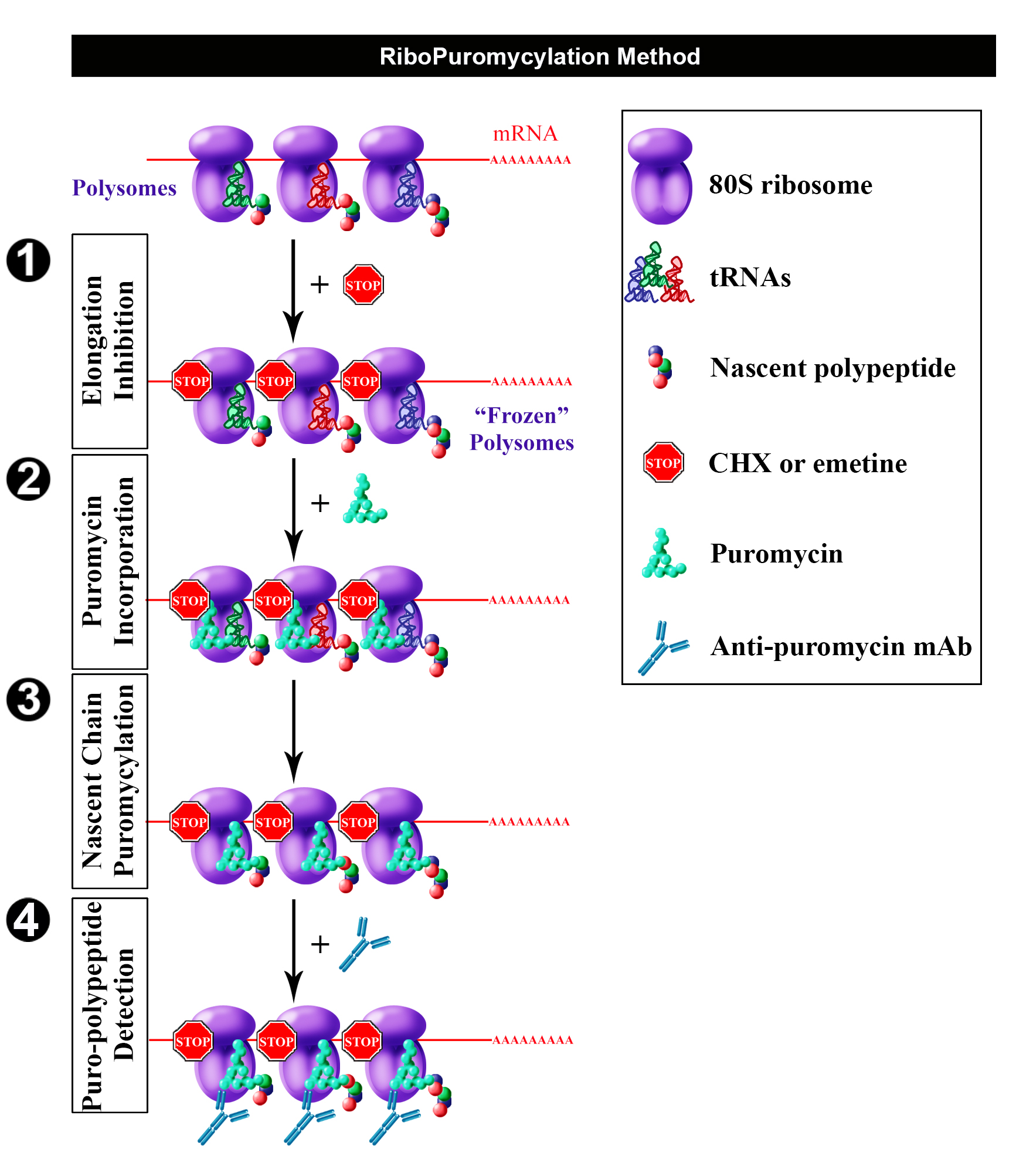
Figure 1. Schematic representation of RPM (from David et al., 2012b). Following freezing of polysome with an elongation inhibitor (step 1), PMY is added (step 2) to living cells and nascent chains become puromycylated through ribosome catalysis (step 3). Anti-PMY monoclonal antibodies detect puromycylated nascent chains via indirect immunofluorescence (step 4). Reproduced from David et al. (2012b) with permission of the publisher and of Dr. Yewdell.
Materials and Reagents
- Materials
- 6-well and 24-well plates (Corning, Costar®, catalog numbers: 3506 , 3524 )
- High quality glass coverslips, 12 mm diameter, #1 thickness (Glaswarenfabrik Karl Hecht, Assistent, catalog number: 1001/12 )
Note: Either autoclave or sterilize with 70% ethanol before use. - Microscope slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: J1800AMNZ )
- Whatman paper (GE Healthcare, catalog number: 3030-917 )
- Parafilm (Bemis, catalog number: 701606 )
- Petri dishes (Corning, catalog number: 430591 )
- Aluminum foil (Sigma-Aldrich, catalog number: Z691569 )
Manufacturer: Heathrow Scientific, catalog number: HD23534A . - Plastic tips
- 6-well and 24-well plates (Corning, Costar®, catalog numbers: 3506 , 3524 )
- Cell line(s)
The described procedure and associated figure (Figure 2) uses HeLa cells (ATCC, catalog number: CCL-2.1 )
Note: Though this procedure was originally designed for HeLa cells, it can be easily adapted for other adherent cell lines (Graber et al., 2013). However, controls with several inhibitors are needed to validate any adjustment (e.g., Digitonin concentration) of any kind. For non-adherent cells, we describe an alternate protocol (Procedure D). - Reagents
- Alcian blue (Sigma-Aldrich, catalog number: A5268 )
- 70% ethanol (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R40135 )
- Hoechst 33258 (Thermo Fisher Scientific, InvitrogenTM, catalog number: H1398 )
- Fluoromount-G (SouthernBiotech, catalog number: 0100-01 )
- Potassium phosphate monobasic (KH2PO4)
- Sodium chloride (NaCl)
- Na2HPO4·7H2O
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 41966029 )
- Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Penicillin/streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 10378016 )
- Fetal bovine serum (FBS) (Eurobio, catalog number: CVFSVF0101 )
- Digitonin (Wako Pure Chemical Industries, catalog number: 043-21376 )
- Tris-HCl pH 7.5 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15567027 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M0250 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Complete Mini EDTA-free protease inhibitor tablets (Sigma-Aldrich, Roche Diagnostics, catalog number: 11836170001 )
- RNase Out (Life Technologies, catalog number: 100000840 ; or Thermo Fisher Scientific, InvitrogenTM, catalog number: 10777019 )
- DEPC treated water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 750023 )
- 16% paraformaldehyde (PFA) (Electron Microscopy Sciences, catalog number: 15710 )
- Sucrose (Sigma-Aldrich, catalog number: 84097 )
- Saponin (Sigma-Aldrich, catalog number: 84510 )
- Glycine (Sigma-Aldrich, catalog number: G7126 )
- Alcian blue (Sigma-Aldrich, catalog number: A5268 )
- Protein synthesis inhibitors (see Table 1)
- Anisomycin (Sigma-Aldrich, catalog number: A9789 )
- Cycloheximide (CHX) (Sigma-Aldrich, catalog number: C7698 )
- Emetine dihydrochloride (Sigma-Aldrich, catalog number: E2375 )
- Harringtonine (Santa Cruz Biotechnology, catalog number: sc-204771 )
- Puromycin (PMY) (Sigma-Aldrich, catalog number: P7255 )
- Sodium arsenite (NaAsO2) (Sigma-Aldrich, catalog number: S7400 )
- Anisomycin (Sigma-Aldrich, catalog number: A9789 )
- Antibodies
- Primary antibodies
- Anti-PMY mouse monoclonal antibodies: We tested 3 mouse mAbs from different hybridoma clones 12D10, 2A4, 5B12. Clone 12D10 was generated by the Pierre laboratory (Schmidt et al., 2009), and is commercially available (Merck, catalog number: MABE343 ). We generated 2A4 and 5B12. 2A4 cells and supernatant are freely available to the scientific community from the Developmental Studies Hybridoma Bank http://dshb.biology.uiowa.edu/PMY-2A4
- Anti-ribosomal P antibody: human polyclonal autoimmune antiserum (from lupus patients) which recognizes three proteins of the 60S ribosomal subunit, RPLP0, P1 and P2 (Immunovision, catalog number: HPO-0100 )
- Anti-lysyl-tRNA synthetase (KRS) antibody: rabbit polyclonal serum which recognizes KRS enzyme (Abcam, catalog number: ab31532 )
- Secondary antibodies
- Donkey anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch, catalog number: 715-545-150 )
- Donkey anti-rabbit Alexa Fluor 594 (Jackson ImmunoResearch, catalog number: 711-585-152 )
- Donkey anti-human Cy5 (Jackson ImmunoResearch, catalog number: 709-175-149 )
- Solutions (see Recipes)
- Anisomycin stock solution (1,000x)
- Cycloheximide (CHX) stock solution (1,000x)
- Emetine dihydrochloride stock solution (1,000x)
- Harringtonine stock solution (1,000x)
- PMY stock solution (1,000x)
- Sodium arsenite stock solution (1,000x)
- Phosphate buffered saline (PBS)
- Growth medium
- Labeling medium
- Labeling control medium
- Extraction buffer
- Wash buffer
- 3% PFA
- Co-extraction/fixation buffer
- Staining buffer (SB)
- Anisomycin stock solution (1,000x)
Equipment
- Ice bucket
- 37 °C water bath (JULABO, model: TW20 )
- Class II laminar flow hood (FASTER, model: SafeFAST Elite )
- Microscopy ultrafine tweezers (Electron Microscopy Sciences, catalog number: 78522-7 )
- PIPETMAN 1-20 µl (Sartorius, model: Proline® Plus, catalog number: 728030 )
- PIPETMAN 20-200 µl (Sartorius, model: Proline® Plus, catalog number: 728060 )
- PIPETMAN 100-1,000 µl (Sartorius, model: Proline® Plus, catalog number: 728070 )
- Slide tray (Glaswarenfabrik Karl Hecht, Assistent, catalog number: 2700/10 )
- Cell Incubator (Heraeus, model: HERACell )
- Centrifuge (Eppendorf, model: 5415 R )
- Microwave
- Microscopy
Laser scanning confocal microscope: Leica TCS SP5 (Leica Microsystems, model: Leica TCS SP5 ) with an HCX PL APO lambda blue 63.0x 1.40 oil UV objective. We used type FF immersion liquid (Cargille-Sacher Laboratories, catalog number: 16212 )
Note: Other comparable systems may be used.
Software
- LAS AF V2.3.1 software (Leica)
- Imaris (Bitplane) and Huygens Essential Software for image deconvolution using the classical maximum likelihood estimation algorithm (V3.6, Scientific Volume Imaging BV, Hilversum, The Netherlands)
- ImageJ (NIH) https://imagej.net/Downloads
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Bastide, A., Yewdell, J. W. and David, A. (2018). The RiboPuromycylation Method (RPM): an Immunofluorescence Technique to Map Translation Sites at the Sub-cellular Level. Bio-protocol 8(1): e2669. DOI: 10.21769/BioProtoc.2669.
- David, A., Dolan, B. P., Hickman, H. D., Knowlton, J. J., Clavarino, G., Pierre, P., Bennink, J. R. and Yewdell, J. W. (2012b). Nuclear translation visualized by ribosome-bound nascent chain puromycylation. JCB 197(1): 45-57.
分类
神经科学 > 细胞机理 > 胞内信号传导
细胞生物学 > 细胞成像 > 固定细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












