- EN - English
- CN - 中文
Construction of a Single Transcriptional Unit for Expression of Cas9 and Single-guide RNAs for Genome Editing in Plants
构建共表达Cas9和sgRNAs的单一独立转录单元用于植物基因组编辑
发布: 2017年09月05日第7卷第17期 DOI: 10.21769/BioProtoc.2546 浏览次数: 13727
评审: Rainer MelzerAlberto CarbonellAnonymous reviewer(s)
Abstract
The CRISPR (clustered regularly interspaced short palindromic repeats)-associated protein9 (Cas9) is a simple and efficient tool for genome editing in many organisms including plant and crop species. The sgRNAs of the CRISPR/Cas9 system are typically expressed from RNA polymerase III promoters, such as U6 and U3. In many transformation events, more nucleotides will increase the difficulties in plasmid construction and the risk of wrong integration in genome such as base-pair or fragment missing (Gheysen et al., 1990). And also, in many organisms, Pol III promoters have not been well characterized, and heterologous Pol III promoters often perform poorly (Sun et al., 2015). Thus, we have developed a method using single transcriptional unit (STU) CRISPR-Cas9 system to drive the expression of both Cas9 and sgRNAs from a single RNA polymerase II promoter to achieve effective genome editing in plants.
Keywords: CRISPR (CRISPR)Background
The sgRNA of the CRISPR-Cas9 system is mainly promoted by the small nuclear RNA promoters such as U6 and U3. Although it has been tested with prospered efficiency in many cases, it also has some limitations: (1) it is hard to achieve coordinated and/or inducible expression of Cas9 and the sgRNAs; (2) manipulating multiple sgRNAs for multiplexed gene editing can be tedious, requiring multiple Pol III promoters. The traditional RNA polymerase II promoter can’t be used in driving sgRNA expression, extra nucleotides will be added to the 5’- and 3’-ends of gRNA by RNA polymerase II and may interrupt the normal gRNA function. Additionally, RNAs transcribed by RNA polymerase II are exported rapidly into the cytoplasm while nuclear localization is required for the CRISPR-Cas9/gRNA duplex to access the genome editing (Lei et al., 2001). To overcome these obstacles, we use the ribozyme’s self-catalyzed cleavage to release the precise processing mature sgRNA under a RNA polymerase II promoter which drives expression of both Cas9 and sgRNA (named STU CRISPR-Cas9 system, Figure 1). Compared to the traditional small nuclear RNA promoters used in sgRNA expression, our STU CRISPR-Cas9 system has some advantages: (1) it’s shorter and easier in vector construction, and it will increase the transformation efficiency under some circumstances; (2) it only needs extra ribozyme flanking sequence (shorter than any RNA polymerase III promoter we are currently using) for multiple sgRNAs expression;(3) it has shown higher deletion efficiency induced by double sgRNAs. Thus, the STU CRISPR-Cas9 system driven by a single RNA polymerase II promoter can replace the traditional CRISPR-Cas9 system now we are using whether in vivo or in vitro if appropriate promoters are chosen.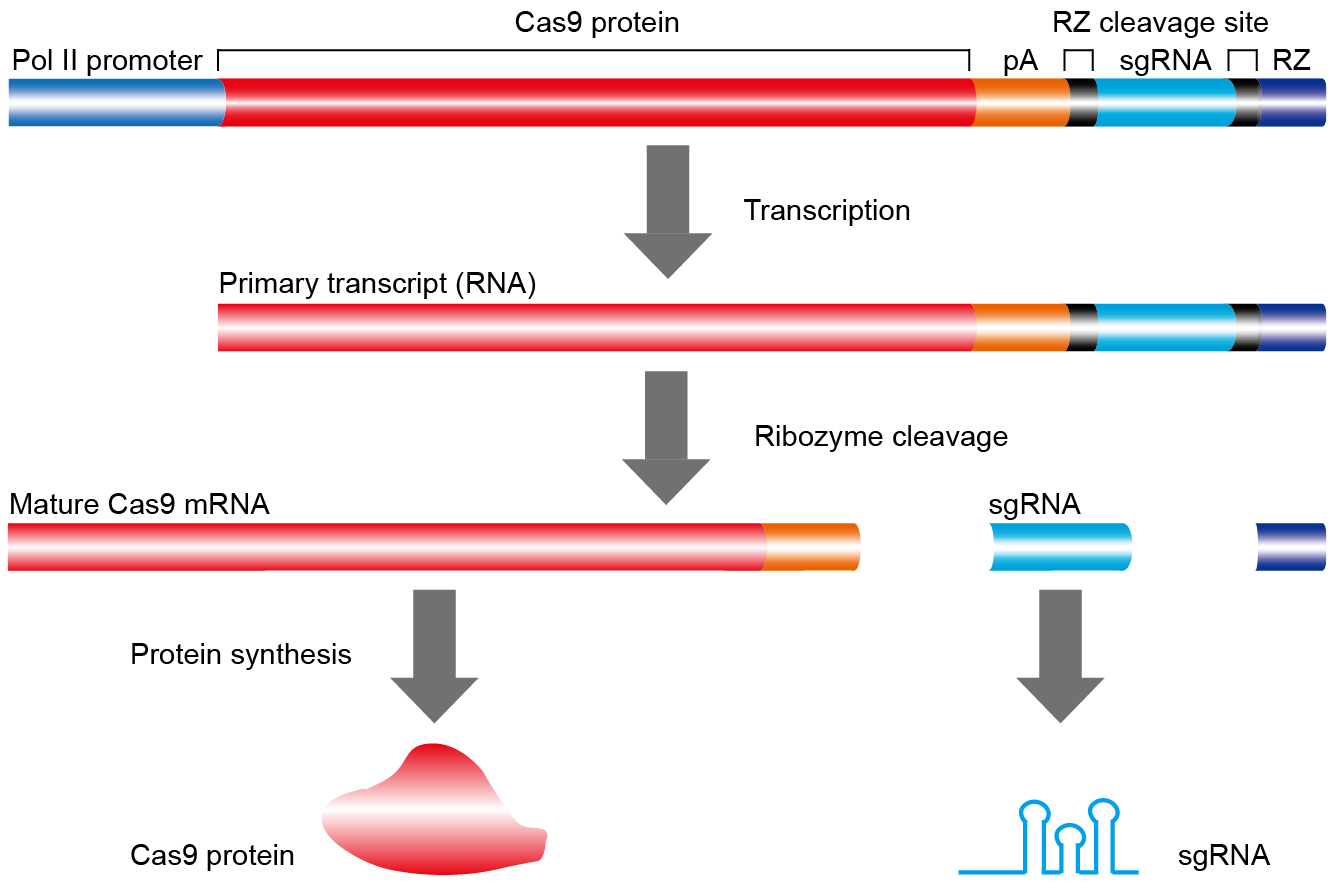
Figure 1. Schematic illustration of the single transcription unit (STU) CRISPR-Cas9 system. Once transcribed by a Pol II promoter, the STU CRISPR-Cas9 primary transcripts will undergo self-cleavage by hammerhead ribozyme (RZ) to release the mature Cas9 mRNA and sgRNA. The Cas9 mRNA is terminated with a synthetic polyA (pA) sequence to facilitate translation, The RZ sequence (in blue) and its target sequence (in black) are illustrated.
Materials and Reagents
- 0.2 ml PCR tubes (Biosharp, catalog number: BS-02-P )
- 1.5 ml Eppendorf tubes (Biosharp, catalog number: BS-15-M )
- Pipette tips (Biosharp, catalog numbers: BS-10-T , BS-200-T , BS-1000-T )
- Competent E. coli DH5α cells (Homemade)
- pTX171 plasmids (Addgene, catalog number: 89258 )
- pTX172 plasmids (Addgene, catalog number: 89259 )
- BsaI (New England Biolabs, catalog number: R0535L )
- Deionized water (sterile)
- Agarose (Biowest, catalog number: 111860 )
- Ethidium bromide (Solarbio Life Scientific, catalog number: E1020 )
- AxyPrepTM DNA Gel Extraction Kit (Corning, Axygen®, catalog number: AP-GX-250 )
- T4 DNA ligase (New England Biolabs, catalog number: M0202L )
- dNTPs mixture (Tiangen Biotech, catalog number: CD117-11 )
- Taq DNA polymerase (Tiangen Biotech, catalog number: ET101-01-02 )
- Q5® High-Fidelity DNA polymerase (New England Biolabs, catalog number: M0491L )
- AxyPrepTM Plasmid Miniprep Kit (Corning, Axygen®, catalog number: AP-MN-P-250 )
- Kanamycin (Solarbio Life Scientific, catalog number: K8020 )
- TAE electrophoresis buffer (see Recipes)
Tris (Solarbio Life Scientific, catalog number: T8060 )
Acetic acid (Kelong)
0.5 M EDTA (Solarbio Life Scientific, catalog number: E1170 )
- LB medium (see Recipes)
Tryptone (Oxoid, catalog number: LP0042 )
Yeast extract (Oxoid, catalog number: LP0021 )
Sodium chloride (NaCl) (Kelong)
Equipment
- Pipettes (Dragon-Lab)
- Heating block (Hangzhou Allsheng Instruments, model: MK-20 )
- Thermal cycler (Thermo Fisher Scientific, Thermo ScientificTM, model: ArktikTM Thermal Cycler )
- Water bath (Yongguangming, model: DZKW-S-4 )
- Microcentrifuge (Eppendorf, model: 5424 )
- DNA electrophoresis apparatus (Bio-Rad Laboratories, model: Mini-Sub® Cell GT Systems )
- NanoDrop (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 )
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Tang, X., Zhong, Z., Zheng, X. and Zhang, Y. (2017). Construction of a Single Transcriptional Unit for Expression of Cas9 and Single-guide RNAs for Genome Editing in Plants. Bio-protocol 7(17): e2546. DOI: 10.21769/BioProtoc.2546.
分类
植物科学 > 植物分子生物学 > DNA > 诱/突变
分子生物学 > DNA > DNA 克隆
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link