- EN - English
- CN - 中文
DNA-free Genome Editing of Chlamydomonas reinhardtii Using CRISPR and Subsequent Mutant Analysis
利用CRISPR技术对莱茵衣藻进行DNA-free的基因组编并对产生突变进行分析
(*contributed equally to this work) 发布: 2017年06月05日第7卷第11期 DOI: 10.21769/BioProtoc.2352 浏览次数: 15981
评审: Vinay PanwarKaisa KajalaAnonymous reviewer(s)
Abstract
We successfully introduced targeted knock-out of gene of interest in Chlamydomonas reinhardtii by using DNA-free CRISPR. In this protocol, the detailed procedures of an entire workflow cover from the initial target selection of CRISPR to the mutant analysis using next generation sequencing (NGS) technology. Furthermore, we introduce a web-based set of tools, named CRISPR RGEN tools (http://www.rgenome.net/), which provides all required tools from CRISPR target design to NGS data analysis.
Keywords: Genome editing (基因组编辑)Background
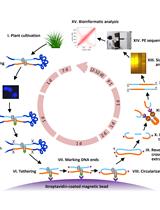
We recently reported (Baek et al., 2016) a one-step transformation of the model green microalga Chlamydomonas reinhardtii (Harris, 2001) using preassembled Cas9 protein-guide RNA ribonucleoproteins (RNPs). The manner of DNA-free CRISPR-Cas9 delivery has several advantages such as no need for codon optimization and specific promoters in different species of microalgae. Furthermore, it reduces off-target effects and may also be less cytotoxic in cells because the Cas9 protein is transiently active and then degraded by endogenous proteases in cells (Kim et al., 2014). In addition, the resulting gene-edited microalgae could be exempt from genetically modified organism (GMO) regulations due to the absence of foreign DNA sequences. In this protocol, the detailed procedures of an entire workflow are contained from the initial target selection of CRISPR to the mutant analysis using NGS technology (Bae et al., 2014a and 2014b; Park et al., 2015 and 2017).
Materials and Reagents
- Cas9 protein purification
- 1.5 ml Eppendorf tubes
- Sterile 50 ml conical tubes
- 10 ml syringe
- 0.45 μm syringe filter
- Protein concentrator–100K MWCO (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 88533 )
- Poly-Prep Chromatography columns (Bio-Rad Laboratories, catalog number: 7311550 )
- BL21-Pro cells
- pET28 plasmid containing 6xHis-SpCas9 (Addgene)
- Kanamycin
- Lysozyme
- PMSF
- Ni-NTA agarose beads (QIAGEN, catalog number: 30210 )
- Bradford reagent (Bio-Rad Laboratories, catalog number: 5000205 )
- Bovine serum albumin (BSA)
- Sodium chloride (NaCl)
- Tryptone
- Yeast extract
- IPTG
- Sodium phosphate monobasic (NaH2PO4)
- Imidazole
- Sodium hydroxide (NaOH)
- HEPES
- Ethylenediaminetetraacetic acid (EDTA)
- DL-dithiothreitol (DTT)
- Sucrose
- Glycerol
- LB medium (see Recipes)
- 1 M IPTG stock (see Recipes)
- Lysis buffer (see Recipes)
- Wash buffer (see Recipes)
- Elution buffer (see Recipes)
- Cas9 storage buffer (see Recipes)
- 1.5 ml Eppendorf tubes
- In vitro transcription and library PCR
- 1.5 ml Eppendorf tubes
- Forward and reverse oligos (see Tables 1 and 2)
Table 1. Pre-index primersPre-index forward primer 5’-ACACTCTTTCCCTACACGACGCTCTTCCGATCT gDNA target-3’ Pre-index reverse primer 5’-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT gDNA target-3’
Table 2. Index primer sequencesindex sequence [i5] Index forward primer sequence D501 tatagcct AATGATACGGCGACCACCGAGATCTACACtatagcctACACTCTTTCCCTACACGAC D502 atagaggc AATGATACGGCGACCACCGAGATCTACACatagaggcACACTCTTTCCCTACACGAC D503 cctatcct AATGATACGGCGACCACCGAGATCTACACcctatcctACACTCTTTCCCTACACGAC D504 ggctctga AATGATACGGCGACCACCGAGATCTACACggctctgaACACTCTTTCCCTACACGAC D505 aggcgaag AATGATACGGCGACCACCGAGATCTACACaggcgaagACACTCTTTCCCTACACGAC D506 taatctta AATGATACGGCGACCACCGAGATCTACACtaatcttaACACTCTTTCCCTACACGAC D507 caggacgt AATGATACGGCGACCACCGAGATCTACACcaggacgtACACTCTTTCCCTACACGAC D508 gtactgac AATGATACGGCGACCACCGAGATCTACACgtactgacACACTCTTTCCCTACACGAC index sequence [i7] Index reverse primer sequence D701 cgagtaat CAAGCAGAAGACGGCATACGAGATcgagtaatGTGACTGGAGTTCAGACGTGT D702 tctccgga CAAGCAGAAGACGGCATACGAGATtctccggaGTGACTGGAGTTCAGACGTGT D703 aatgagcg CAAGCAGAAGACGGCATACGAGATaatgagcgGTGACTGGAGTTCAGACGTGT D704 ggaatctc CAAGCAGAAGACGGCATACGAGATggaatctcGTGACTGGAGTTCAGACGTGT D705 ttctgaat CAAGCAGAAGACGGCATACGAGATttctgaatGTGACTGGAGTTCAGACGTGT D706 acgaattc CAAGCAGAAGACGGCATACGAGATacgaattcGTGACTGGAGTTCAGACGTGT D707 agcttcag CAAGCAGAAGACGGCATACGAGATagcttcagGTGACTGGAGTTCAGACGTGT D708 gcgcatta CAAGCAGAAGACGGCATACGAGATgcgcattaGTGACTGGAGTTCAGACGTGT D709 catagccg CAAGCAGAAGACGGCATACGAGATcatagccgGTGACTGGAGTTCAGACGTGT D710 ttcgcgga CAAGCAGAAGACGGCATACGAGATttcgcggaGTGACTGGAGTTCAGACGTGT D711 gcgcgaga CAAGCAGAAGACGGCATACGAGATgcgcgagaGTGACTGGAGTTCAGACGTGT D712 ctatcgct CAAGCAGAAGACGGCATACGAGATctatcgctGTGACTGGAGTTCAGACGTGT - dNTP mix
- Phusion DNA polymerase (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: F530L )
- PCR purification kit
- ATP, CTP, GTP, UTP, 100 mM MgCl2, DEPC-treated water
- T7 RNA polymerase (New England Biolabs, catalog number: M0251L )
- DNase I (RNase-free) (New England Biolabs, catalog number: M0303L )
- RNase inhibitor murine (New England Biolabs, catalog number: M0314L )
- RNeasy MinElute Cleanup Kit (QIAGEN, catalog number: 74204 )
- Illumina Miseq Reagent Kit (v2)
- 1.5 ml Eppendorf tubes
- Transfection
- 1.5 ml Eppendorf tubes
- Sterile 15 ml and 50 ml conical tubes
- Purified Cas9 protein and sgRNA
- Chlamydomonas reinhardtii strains CC-4349 cw15 mt-(Chlamydomonas Resource Center)
- PCR DNA purification kit (MG Med, catalog number: MD008 )
- TAP medium (Harris, 1989 or Thermo Fisher Scientific, GibcoTM, catalog number: A1379801 )
- TAP sucrose solution (see Recipes)
- TAP agar plates (see Recipes)
- Top agar (see Recipes)
- 1.5 ml Eppendorf tubes
Equipment
- Cas9 protein purification
- In vitro transcription and library PCR
- Incubator (JS Research, model: JSGI-050T )
- Thermal cycler (Bio-Rad Laboratories, model: C1000 TouchTM Thermal Cycler )
- Micro centrifuge (LaboGene, model: 1730R )
- Spectrophotometer
- Incubator (JS Research, model: JSGI-050T )
- Transfection
- 100 ml flask
- Spectrophotometer (GE Healthcare, Amersham Biosciences, model: Ultrospec 2100 pro )
- Hemocytometer (Marienfeld-Superior, catalog number : 0650030 )
- Microscope (Olympus, model: CH30 )
- Micro high speed centrifuge (Hanil, model: MICRO 17TR )
- Orbital shaker (N-biotek, model: NB-101M )
- Clean bench (BioFree, model: BF-150BSC )
- Electroporation device (Bio-Rad Laboratories, model: Gene Pulser XcellTM Electroporation Systems )
- Electroporation cuvettes (Bio-Rad Laboratories, 0.4 cm gap)
- 100 ml flask
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Yu, J., Baek, K., Jin, E. and Bae, S. (2017). DNA-free Genome Editing of Chlamydomonas reinhardtii Using CRISPR and Subsequent Mutant Analysis. Bio-protocol 7(11): e2352. DOI: 10.21769/BioProtoc.2352.
分类
植物科学 > 植物转化 > 电穿孔
细胞生物学 > 细胞工程 > CRISPR-cas9
植物科学 > 植物分子生物学 > DNA > DNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link