- EN - English
- CN - 中文
Targeted Nucleotide Substitution in Mammalian Cell by Target-AID
在哺乳动物细胞中通过靶向性AID进行核苷酸靶向置换
发布: 2017年06月05日第7卷第11期 DOI: 10.21769/BioProtoc.2339 浏览次数: 19738
评审: Daan C. SwartsZhen ShiAnonymous reviewer(s)

相关实验方案

利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1216 阅读
Abstract
Programmable RNA-guided nucleases based on CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated protein) systems have been applied to various type of cells as powerful genome editing tools. By using activation-induced cytidine deaminase (AID) in place of the nuclease activity of the CRISPR/Cas9 system, we have developed a genome editing tool for targeted nucleotide substitution (C to T or G to A) without donor DNA template (Figure 1; Nishida et al., 2016). Here we describe the detailed method for Target-AID to perform programmable point mutagenesis in the genome of mammalian cells. A specific method for targeting the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene in Chinese Hamster Ovary (CHO) cell was described here as an example, while this method principally should be applicable to any gene of interest in a wide range of cell types.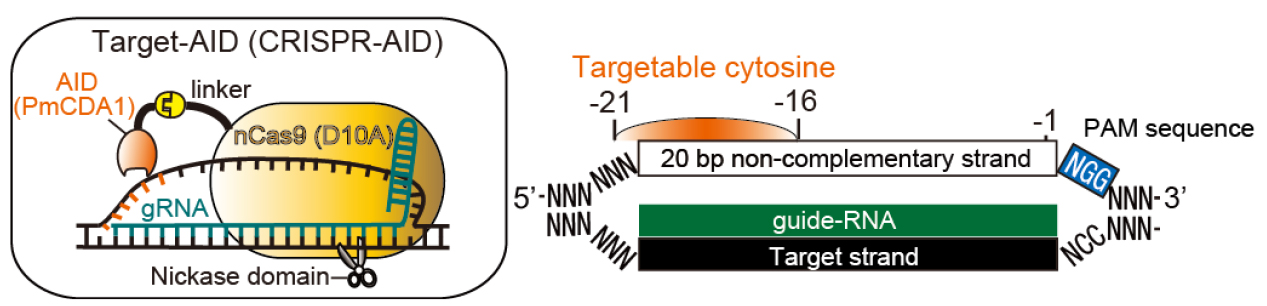
Figure 1. Schematic illustration for Target-AID and its targetable site. In a guide-RNA (gRNA)-dependent manner, PmCDA1 fused to nCas9 (D10A) via a linker performs programmable cytidine mutagenesis around -21 to -16 positions relative to PAM sequence on the non-complementary strand in mammalian cells. The targetable site was determined based on the efficient base substitution (> 20%) observed in the previous work.
Background
Insertion or deletion caused by DNA double strand break at the target site is efficiently induced to disrupt gene function. However, more precise genome modifications are still limited as homology directed repair is not always efficient enough in higher eukaryotes, especially when considering delivery of template DNA for in vivo genome editing. In addition, CRISPR nucleases also have some potential for off-target effect by cutting the genome (Cox et al., 2015). Target-AID demonstrated a very narrow range of targeted nucleotide modification without use of template DNA. AID can convert cytosine to uracil without DNA cleavage by deamination and then, uracil is converted to thymine or the other bases through DNA replication and/or repair. Use of uracil DNA-glycosylase inhibitor (UGI), which blocks removal of uracil in DNA and the subsequent repair pathway, rendered mutations more likely to be C to T substitutions and improved the efficiency. While a series of variable components for Target-AID had been tested such as linkage, nickase Cas9 (nCas9) and UGI in the original study, we will focus on the use of AID ortholog PmCDA1 derived from sea lamprey, fused to nCas9 or nCas9 plus UGI for simplicity. Consistent to our study, applying the rat apolipoprotein B mRNA editing enzyme, catalytic polypeptide (rAPOBEC1) has also been reported as a programmable base editor (BE). Although BE targeted 5 bases surrounding the -15 position upstream of PAM (Komor et al., 2016), Target-AID can modify 3 to 6 bases surrounding the -18 position upstream PAM. More recently, it has been reported that Target-AID can be applied for precise editing of plant genome (Shimatani et al., 2017).
Materials and Reagents
- Cell culture-treated polystyrene 24 well plate (Sumitomo Bakelite, catalog number: MS-80240Z )
- 100 mm dish (TPP, catalog number: 93100 )
- 15 ml and 1.5 ml tubes
- 200 μl pipette tips
- CHO-K1 cells (ECACC, catalog number: 85051005 )
- Target-AID vectors
nCas9-PmCDA1 (Addgene, catalog number: 79617 )
nCas9-PmCDA1-UGI (Addgene, catalog number: 79620 ) - Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070 )
- Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668019 )
- Dulbecco’s phosphate buffered saline (D-PBS) (Nacalai Tesque, catalog number: 14249-24 )
- NucleoSpin Tissue XS (MACHEREY-NAGEL, catalog number: 740901.50 )
- A pair of primers to amplify the target genomic region plus 150-200 bp upstream and downstream sequences (for HPRT target1, Fw: 5’-GGCTACATAGAGGGATCCTGTGTCA-3’; Rev: 5’-ACAGTAGCTCTTCAGTCTGATAAAA-3’) (Eurofin genomics)
- KOD FX Neo (TOYOBO, catalog number: KFX-201 )
- Gel extraction kit (QIAGEN, catalog number: 28704 )
- (Optional) NEBNext Multiplex Oligos for Illumina (Dual Index Primer Set1) (New England Biolabs, catalog number: E7600S )
- (Optional) MiSeq reagent Kit v3 (Illumina, catalog number: MS-102-3003 )
- Ham’s F12 medium (Thermo Fisher Scientific, GibcoTM, catalog number: 11765054 )
- Fetal bovine serum (FBS) (Biosera, catalog number: FB-1360/500 )
- Penicillin-streptomycin (Nacalai Tesque, catalog number: 26253-84 )
- G418
- Trypsin-EDTA, 0.25% (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056 )
- (Optional) 6-TG
- Ham’s F12 culture medium (see Recipes)
- Ham’s F12-G418 culture medium (see Recipes)
- Trypsin-EDTA 0.025% (see Recipes)
- (Optional) Ham’s F12-G418-6-TG culture medium (see Recipes)
Equipment
- Cell culture incubator at 37 °C with 5% CO2 (Panasonic, catalog number: KMCC17RU2J ) or equivalent
- Micropipette
- Optical microscope with 10x eyepiece and 10x objective lens (Olympus, model: CKX41 ) or equivalent
- Cell counting plate (WAKENBTECH, catalog number: OC-C-S02 )
- Centrifuge (Max speed: 15,000 rpm; Max RCF: 21,380 x g; 24 x 1.5/2.0 ml angle rotor; 12 x 15 ml swing-out rotor) (KUBOTA, model: 3740 ) or equivalent
- Heat block (TAITEC, model: CTU-Mini , catalog number: 0063288-000) or equivalent
- PCR thermal cycler (TaKaRa Bio, model: TP600 ) or equivalent
- Agarose gel electrophoresis system
- 3130xL Genetic Analyzer (Thermo Fisher Scientific, Applied BiosystemsTM, model: 3130xL Genetic Analyzer ) or equivalent
Software
- CLC Genomic workbench 7.0
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Arazoe, T., Nishida, K. and Kondo, A. (2017). Targeted Nucleotide Substitution in Mammalian Cell by Target-AID. Bio-protocol 7(11): e2339. DOI: 10.21769/BioProtoc.2339.
分类
癌症生物学 > 通用技术 > 遗传学 > DNA损伤
细胞生物学 > 细胞工程 > CRISPR-cas9
分子生物学 > DNA > DNA 损伤和修复
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link