- EN - English
- CN - 中文
Generation of Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors
通过直接原核显微注射CRISPR/Cas9质粒载体生成突变株
发布: 2017年06月05日第7卷第11期 DOI: 10.21769/BioProtoc.2321 浏览次数: 13052
评审: Longping Victor TseFang XuAnonymous reviewer(s)

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1216 阅读
Abstract
A set of Cas9 and single guide CRISPR RNA expression vectors was constructed. Only a very simple procedure was needed to prepare specific single-guide RNA expression vectors with high target accuracy. Since the de novo zygotic transcription had been detected in mouse embryo at the 1-cell stage, the plasmid DNA vectors encoding Cas9 and GGTA1 gene specific single-guide RNAs were micro-injected into zygotic pronuclei to confirm such phenomenon in 1-cell pig embryo. Our results demonstrated that mutations caused by these CRISPR/Cas9 plasmids occurred before and at the 2-cell stage of pig embryos, indicating that besides the cytoplasmic microinjection of in vitro transcribed RNA, the pronuclear microinjection of CRISPR/Cas9 DNA vectors provided an efficient solution to generate gene-knockout pig.
Keywords: CRISPR/Cas9 (CRISPR/Cas9)Background
Since the initial discovery of a highly conserved 29 base pairs (bp) sequence tandemly repeated with a spacing of 32 bp downstream of the iap gene in Escherichia coli genome (Ishino et al., 1987; Nakata et al., 1989), a family of short regularly spaced repeats, varying in size from 25 to 50 bp, were found in about 50% of bacteria and 90% of archaea (Makarova et al., 2015). According to their characteristic structures, the name clustered regularly interspaced short palindromic repeats (CRISPRs) were introduced by Mojica (Mojica et al., 2009) and Jansen (Jansen et al., 2002) and are currently in general use. A set of CRISPR associated genes, cas1 to cas4, was first identified flanking the CRISPR loci by nucleotide sequence alignments (Jansen et al., 2002). New members of cas genes were identified in different bacterial species. According to the cas members within each host, the CRISPR-Cas systems can be classified into three major types based on the hallmark genes (Makarova et al., 2011; Burmistrz and Pyrc, 2015). The function of the CRISPR-Cas system was demonstrated by a seminar experiment. After being challenged by virulent bacteriophages: phage 858 and phage 2,972, new repeat-spacer units were observed on the leading end of the CRISPR array in the surviving Streptococcus thermophlis host cells. The DNA sequences of newly acquired spacers were matched to corresponding fragments, named proto-spacers, in the phage genomes. Streptococcus thermophlis strains with phage spacer(s) were resistant to the phage infection, while strains without phage spacer(s) were sensitive to the phage infection (Barrangou et al., 2007). To distinguish between the spacer in the host bacterial genome and the proto-spacer, which has the same sequence as the spacer in the invader genome, a proto-spacer adjacent motif (PAM) was evolved (Mojica et al., 2009; Shah et al., 2013). The CRISPR-Cas immunity was revealed, it can be divided into three stages (Rath et al., 2015; Wright et al., 2016). The first adaptation or acquisition stage is responsible for the acquisition of spacers into CRISPR array following the exposure to foreign mobile genetic elements, such as phages or plasmids. A Cas2 homodimer was sandwiched by two Cas1 homodimers forming a heterohexameric complex for all three types of CRISPR-Cas systems (Sternberg et al., 2016). In the second stage, the promoter embedded within the AT-rich leader sequence upstream of the CRISPR array transcribed the precursor CRISPR RNAs (pre-crRNAs), these were further processed into short CRISPR RNA (crRNA) guides by Cas proteins. Cas6 was involved in the RNA processing step in both of the type I and type III CRISPR-Cas systems (Charpentier et al., 2015; Hochstrasser and Doudna, 2015). Accompanied by the Cas9 protein, a trans-activating crRNA (tracrRNA) which contains an anti-repeat segment for duplex formation with the repeat compartment of crRNA was involved in the maturation of the crRNAs in the type II system (Deltcheva et al., 2011). In the last interference stage, in cooperation with a mature crRNA and a cascade of Cas proteins, the signature proteins Cas3 and Cas10 were integrated into the RNA guided endonuclease complex in the type I and type III CRISPR-Cas systems, respectively. The type II effector is simply composed of a Cas9 protein, a pair of processed mature crRNA and tracrRNA (Gasiunas et al., 2012). The crRNA and tracrRNA in the ternary complex can be substituted by a fused crRNA-tracrRNA single-guide RNA (sgRNA) (Jinek et al., 2012). Because of extreme simplicity, the Cas9-sgRNA, now commonly termed as CRISPR/Cas9, binary complexes were immediately applied in the field of gene editing (Cong et al., 2013; Mali et al., 2013).
Swine share a number of anatomic and physiologic characteristics with humans. Systems that are mostly cited as suitable models include cardiovascular, urinary, integumentary, and digestive system (Swindle et al., 2012). Therefore, pigs are considered as a good source of organs for xenotransplantation. Two strategies are currently utilized to overcome the interspecies rejection hurdles. The first one is to block donor organs expressing the antigens causing hyper-acute rejection, such as galactose-α1,3-galactose, N-glycolylneuroaminic acid and β1,4-N-acetylgalactosamine, by targeting the GGTA1, CMAH and β4GalNT2 genes, respectively (Estrada et al., 2015; Cooper et al., 2016). The second strategy is to prepare organs which are composed of acceptor’s cells in a surrogate animal by a blastocyst complementation technique (Kobayashi et al., 2010; Usui et al., 2012; Matsunari et al., 2013). To produce gene-knockout pigs, the traditional method uses gene editing in somatic cells, such as fetal fibroblasts, and somatic cell nuclear transfer (SCNT) techniques to create knockout zygotes. The examples of combining CRISPR/Cas9 and SCNT are reported to generate IgM JH (Chen et al., 2015), RUNX3 (Kang et al., 2016b), IL2RG (Kang et al., 2016a), and GGTA1/CMAH/β4GalNT2 triple knockout pigs (Estrada et al., 2015). Direct microinjection of DNA or RNA is another choice to prepare gene knockout pigs. Since the burst of de novo transcription in porcine zygotes was reported at the 4-cell stage (Anderson et al., 1999), RNA is preferred for micro-injection into embryos at the 1-cell stage. Previous studies have demonstrated that cytoplasmic microinjection of Cas9 mRNA with sgRNAs can produce Mitf (Wang et al., 2015) and DJ-1/Parkin/PINK1 triple-gene knockout pigs (Wang et al., 2016). Because de novo zygotic transcription had only been reported in the 1-cell stage mouse embryo (Ram and Schultz, 1993; Bouniol et al., 1995; Aoki et al., 1997), a trial was reported to produce GGTA1 knockout pigs by cytoplasmic microinjection of a CRISPR/Cas9 plasmid. With this strategy, CRISPR/Cas9 was expressed at, or later, than the 2-cell stage, and mosaic mutations on GGTA1 gene were found (Petersen et al., 2016). Pronucleus microinjection is needed to interpret whether zygotic transcription occurs at the 1-cell stage of pig embryo (Chuang et al., 2016).
A series of Cas9 and sgRNA expression vectors was constructed as shown in Figure 1. pCX-Flag2-NLS1-Cas9-NLS2, pCX-HA-NLS1-Cas9-NLS2, and pCX-Myc-NLS1-Cas9-NLS2 can be used to express Cas9 in mammalian cells. Three common tags are available to monitor Cas9 expressions. (Figure 1A) Porcine U6 promoter which could effectively derive short hairpin RNA [Chuang et al., 2009] was used to construct the ppU6-(BsaI)2-gRNA vector (Figure 1B) (Su et al., 2015). A pair of primers containing spacer and part of CRISPR repeat sequences, as shown in Figure 1B, is needed for each target site. Because guanine (G) is favored for U6 promoter as the first transcribed nucleotide, only proto-spacers initiated with G were chosen in our recent works.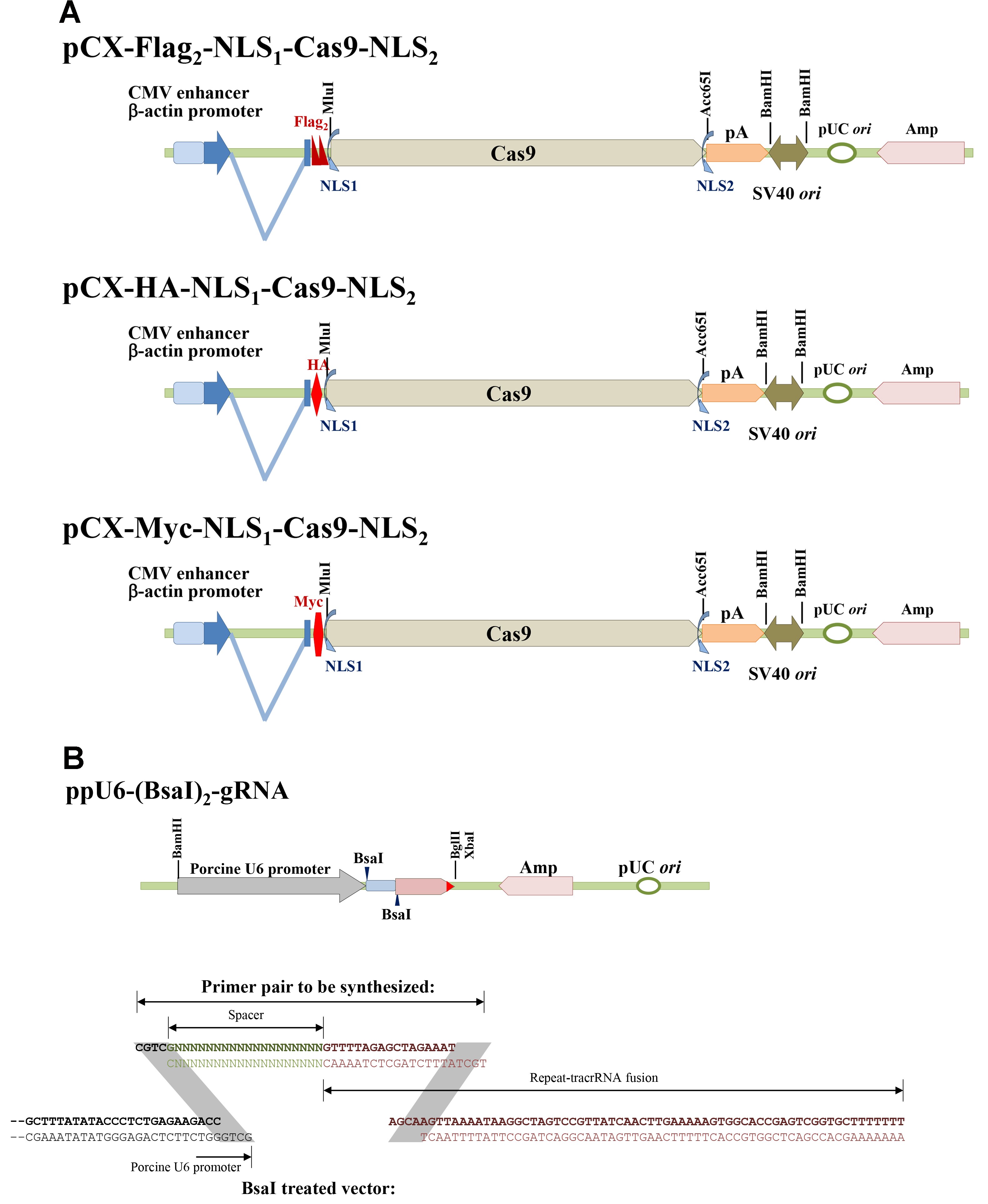
Figure 1. Scheme of the Cas9 and single-guide RNA expression vectors. A. pCX-Flag2-NLS1-Cas9-NLS2, pCX-HA-NLS1-Cas9-NLS2, and pCX-Myc-NLS1-Cas9-NLS2; B. ppU6-(BsaI)2-gRNA and primer pair designation.
Materials and Reagents
- Preparations of sgRNA expression vectors
- BsaI (New England Biolabs, catalog number: R0535 )
- T4 DNA ligase (Promega, catalog number: M1801 )
- Clean and Gel Extraction Kit (Biokit, catalog number: Bio-C300 )
- 2x ligation buffer (Promega, catalog number: C671A )
- pGEM-T Easy TA-cloning kit (Promega, catalog number: A1360 )
- Plasmid Miniprep Kit (Biokit, catalog number: Bio-P300 )
- Genomic DNA isolation Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: K0721 )
- NeonTM Transfection System 100 µl Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: MPK10096 )
- Cesium chloride (CsCl) (Avantor® Performance Materials, J.T. Baker®, catalog number: 4042-02 )
- Tris-HCl, pH 8.0
- EDTA, pH 8.0
- Sodium chloride (NaCl)
- TE buffer (see Recipes)
- TEN buffer (see Recipes)
- BsaI (New England Biolabs, catalog number: R0535 )
- Collection of pig embryos
- Glass tube for embryo flashing (glass tube with outside diameter of 4 mm as shown in Figure 2)
- Falcon tube, 50 ml (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339653 )
- Regumate® (containing 0.4% Altrenogest, Intervet, MSD, France, brand name: Regumate®)
- PMSG (pregnant mare serum gonadotropin) (ASFK Pharmaceutical, Japan, brand name: PMSG)
- hCG (human chorionic gonadotropin) (ASKA Pharmaceutical, Japan, brand name: hCG)
- PGF2α (prostaglandin F2α; Estrumate injection) (Intervet Deutschland, Germany)
- D-PBS (GE Healthcare, HyCloneTM, catalog number: SH30028.02 )
- Fetal bovine serum (FBS) (GE Healthcare, HyCloneTM, catalog number: SH30071.03 )
- Glass tube for embryo flashing (glass tube with outside diameter of 4 mm as shown in Figure 2)
- Embryo transfer
- 3M paper tape (3M, catalog number: 200-24mm )
- Atropine sulfate (TAI YU CHEMICAL & PHARMACEUTICAL)
- Stresnil (azaperona, 40 mg/ml) (Janssen Pharmaceutica N.V., Belgium, brand name: Stresnil)
- Citosol (Thiamylal sodium) (Shinlin Sinseng Pharmaceutical, Taiwan, brand name: Citosol)
- Amoxicillin (ampicillin, 150 mg/ml) (China Chemical & Pharmaceutical, CCPG, catalog number: E000853 )
- Heparin sodium (China Chemical & Pharmaceutical, CCPG, Taiwan, brand name: AGGLUTEX INJECTION)
- 3M paper tape (3M, catalog number: 200-24mm )
Equipment
- Preparations of sgRNA erxpression vectors
- Ultracentrifugator (Beckman Coulter, model: Optima XL-80 ) equipped with NVTi65 rotor
- Microcentrifugator (KUBOTA, model: 3740 )
- Dry bath incubator (Major Science, model: MD-01N )
- Thermocycler (Thermo Fisher Scientific, Applied BiosystemsTM, model: 2720 )
- Ultracentrifugator (Beckman Coulter, model: Optima XL-80 ) equipped with NVTi65 rotor
- Semen assessment
- computer-assisted sperm analysis system (UltiMate CASA system, Hamilton-Thorne Research, Beverly, MA)
- computer-assisted sperm analysis system (UltiMate CASA system, Hamilton-Thorne Research, Beverly, MA)
- Microinjection
- Centrifugator (KUBOTA, model: 8920 )
Note: This product has been discontinued. - Microcentrifugator (KUBOTA, model: 3740 )
- Inverted Differential Interference Contrast microscope (Olympus, model: IX71 )
- Capillary injection needle (Sutter Instrument, catalog number: BF100-78-10 )
- Capillary injection needle puller (Sutter Instrument, model: P-97 )
- Micromanipulator (NARISHGE, model: ON3-99D )
- Injectors (NARISHGE, model: IM-9B )
- Centrifugator (KUBOTA, model: 8920 )
- Collection of pig embryos and embryo transfer
- Operation table (NEWPORT, LW3048B-OPT, VIBRATION ISOLATION TABLE)
- Surgery mechanic devises (NARISHIGE, JAPAN)
- Operation table (NEWPORT, LW3048B-OPT, VIBRATION ISOLATION TABLE)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Chuang, C., Tu, C. and Chen, C. (2017). Generation of Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors. Bio-protocol 7(11): e2321. DOI: 10.21769/BioProtoc.2321.
分类
细胞生物学 > 细胞工程 > CRISPR-cas9
分子生物学 > DNA > 转染
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link