- EN - English
- CN - 中文
CRISPR-PCS Protocol for Chromosome Splitting and Splitting Event Detection in Saccharomyces cerevisiae
检测酿酒酵母中染色体分裂和分裂事件的CRISPR-PCS实验方案
发布: 2017年05月20日第7卷第10期 DOI: 10.21769/BioProtoc.2306 浏览次数: 10967
评审: Renate WeizbauerJudd F HultquistAnonymous reviewer(s)
Abstract
Chromosome engineering is an important technology with applications in basic biology and biotechnology. Chromosome splitting technology called PCS (PCR-mediated Chromosome Splitting) has already been developed as a fundamental chromosome engineering technology in the budding yeast. However, the splitting efficiency of PCS technology is not high enough to achieve multiple splitting at a time. This protocol describes a procedure for achieving simultaneous and multiple chromosome splits in the budding yeast Saccharomyces cerevisiae by a new technology called CRISPR-PCS. At least four independent sites in the genome can be split by one transformation. Total time and labor for obtaining a multiple split yeast strain is drastically reduced when compared with conventional PCS technology.
Keywords: Saccharomyces cerevisiae (酿酒酵母)Background
Chromosome engineering technologies that enable rapid and efficient manipulation of multiple genetic loci or chromosomal regions have become increasingly important. Such technologies offer a powerful means for elucidating chromosome and genome function. Additionally, it can be used for breeding useful strains through the creation of a wide array of genetic variants. A chromosome splitting technology called PCS (PCR-mediated Chromosome Splitting) technology has been developed in the budding yeast Saccharomyces cerevisiae. This technology allows the splitting of yeast chromosomes at any desired site by introducing centromeres and telomere seed sequences based on the homologous recombination mechanism. The resulting chromosomes possess one centromere and telomeres at both ends, thus function as normal chromosomes (Sugiyama et al., 2005). However, low splitting efficiency is a drawback in PCS, therefore simultaneous and multiple splitting of chromosomes has been impossible. In this situation, we developed a novel chromosome splitting technology called CRISPR-PCS. It is well known that double strand break (DSB) markedly increases homologous recombination activity around the DSB site in yeast (Agmon et al., 2009). The CRISPR/Cas9 system is a genome editing technology that can induce targeted DSBs. By utilizing CRISPR/Cas9 system, we can induce DSB at any genomic locus and thus activate homologous recombination activity. CRISPR-PCS is a technology that combines CRISPR/Cas9 system with PCS, thus allowing the increase of splitting efficiency by approximately 200 fold. This drastically increased efficiency enables simultaneous and multiple chromosome spitting. Overview of the CRISPR-PCS technology is illustrated in Figure 1.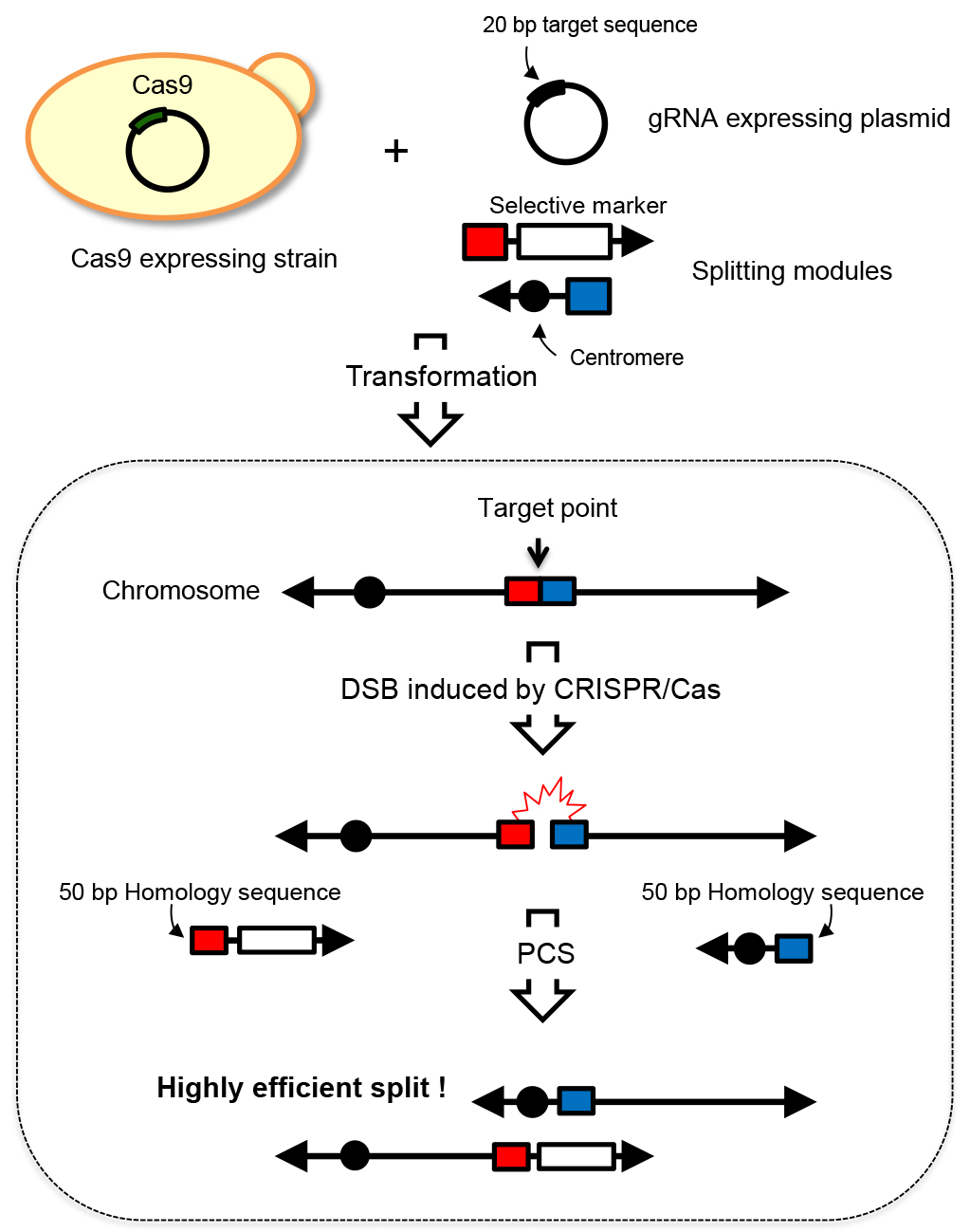
Figure 1. Overview of CRISPR-PCS. In CRISPR-PCS, one gRNA expressing plasmid for the specific targeting site and two splitting modules are required to split yeast chromosome at a specific site. These DNA molecules are introduced into the Cas9 expressing strain, i.e., the strain carrying p414-TEF1p-Cas9-CYC1t plasmid. Transformants where the expected split event occurred are selected by auxotrophic marker selection. Closed black circles represent the centromere. Red and blue boxes represent the homology sequences for recombination. Arrows represent the telomere sequence.
Materials and Reagents
- 10-100 μl pipette tips (e.g., Greiner Bio One International, catalog number: 685280 )
- 100-1,000 μl pipette tips (e.g., Greiner Bio One International, catalog number: 686290 )
- PCR tube (e.g., Greiner Bio One International, catalog number: 683201 )
- p426-SNR52p-gRNA.CAN1.Y-SUP4t (Addgene, catalog number: 43803 )
- p414-TEF1p-Cas9-CYC1t (Addgene, catalog number: 43802 )
- Escherichia coli DH5α competent cell (NIPPON GENE, catalog number: 316-06233 )
- DNA, MB-grade from fish sperm (Roche Diagnostics, catalog number: 11467140001 )
- KOD plus neo (TOYOBO, catalog number: KOD-401 )
- 2 mM dNTP solution (Attached in the KOD plus neo)
- 25 mM magnesium sulfate (MgSO4) (Attached in the KOD plus neo)
- Oligonucleotide primer 1 for construction of a gRNA expressing plasmid (5’-N20GTTTTAGAGCTAGAAATAGCAAG-3’) (synthesized in Sigma-Aldrich Japan)
- Oligonucleotide primer 2 for construction of gRNA expressing plasmid (5’-cN20GATCATTTATCTTTCACTGCGGA-3’) (synthesized in Sigma-Aldrich Japan)
- DpnI (Takara Bio, catalog number: 1235A )
- QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704 )
- 10x NEB buffer 2 (New England Biolabs, catalog number: B7002S )
- BSA solution (attached in the T4 DNA polymerase) (New England Biolabs, catalog number: M0203 )
- 10x T4 DNA ligase buffer (New England Biolabs, catalog number: M0202 )
- T4 DNA polymerase (New England Biolabs, catalog number: M0203 )
- 2.5 mM each dNTP mix (Takara Bio, catalog number: 4030 )
- Ampicillin (Wako Pure Chemical Industries, catalog number: 015-10382 )
- Oligonucleotide primer 1 for construction of a splitting module (5’-N50GGCCGCCAGCTGAAGCTTCG-3’) (synthesized in Sigma-Aldrich Japan)
- Oligonucleotide primer 2 for construction of a splitting module (5’-CCCCAACCCCAACCCCAACCCCAACCCCAACCCCAAAGGCCACTAGTGGATCTGAT-3’) (synthesized in Sigma-Aldrich Japan)
- Oligonucleotide primer for sequencing (5’-ACGCCAAGCGCGCAATTAAC-3’) (synthesized in Sigma-Aldrich Japan)
- Lithium acetate dihydrate (Wako Pure Chemical Industries, catalog number: 120-01535 )
- Polyethylene glycol 4,000 (Wako Pure Chemical Industries, catalog number: 162-09115 )
- ECL Direct Nucleic Acid Labelling and Detection System (GE Healthcare, catalog number: RPN3000 )
- Ethidium bromide solution (10 mg/ml) (Nacalai Tesque, catalog number: 14631-94 )
- LB broth (Sigma-Aldrich, catalog number: L3022-1KG )
- Agar (Wako Pure Chemical Industries, catalog number: 010-08725 )
- Glucose (Wako Pure Chemical Industries, catalog number: 043-31163 )
- Yeast nitrogen base without amino acids (e.g., BD, Difco, catalog number: 291940 )
- Sodium hydroxide (NaOH) (e.g., Wako Pure Chemical Industries, catalog number: 192-15985 )
- Peptone (e.g., BD, BactoTM, catalog number: 211677 )
- Yeast extract (e.g., BD, BactoTM, catalog number: 288620 )
- Hydrochloric acid (HCl) (e.g., Wako Pure Chemical Industries, catalog number: 087-10361 )
- LB plate (see Recipes)
- Yeast minimum medium (SD medium) (see Recipes)
- YPD medium (see Recipes)
Equipment
- Pipettes
- Thermal cycler (e.g., Takara Bio, model: Dice® Touch, catalog number: TP350 ) (Use at Procedure B and Procedure C)
- Incubator (e.g., Panasonic Healthcare, model: MIR-H163 ) (Use at Procedure B and Procedure D)
- Air shaker (e.g., TAITEC, model: BR-21UM MR ) (Use at Procedure B and Procedure D)
- Heat block (e.g., TAITEC, model: DTU-1BN ) (Use at Procedure B, Procedure D and Procedure E)
- DNA sequencer (e.g., Applied Biosystems, model: ABI PRISM® 3100 Genetic Analyzer ) (Use at Procedure B)
- CHEF-DR® III pulsed field gel electrophoresis system (Bio-Rad Laboratories, model: CHEF-DR III Chiller System , catalog number: 1703700)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sasano, Y. and Harashima, S. (2017). CRISPR-PCS Protocol for Chromosome Splitting and Splitting Event Detection in Saccharomyces cerevisiae. Bio-protocol 7(10): e2306. DOI: 10.21769/BioProtoc.2306.
分类
分子生物学 > DNA > 染色体工程
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link















