- EN - English
- CN - 中文
Semi-quantitative Analysis of H4K20me1 Levels in Living Cells Using Mintbody
使用Mintbody半定量分析活细胞中H4K20me1的水平
发布: 2017年05月20日第7卷第10期 DOI: 10.21769/BioProtoc.2276 浏览次数: 9969
评审: Ralph BottcherCody KimeMartin V Kolev

相关实验方案

采用 Davidson 固定液和黑色素漂白法优化小鼠眼组织切片的免疫组化染色
Anne Nathalie Longakit [...] Catherine D. Van Raamsdonk
2025年11月20日 1572 阅读
Abstract
Eukaryotic nuclear DNA wraps around histone proteins to form a nucleosome, a basic unit of chromatin. Posttranslational modification of histones plays an important role in gene regulation and chromosome duplication. Some modifications are quite stable to be an epigenetic memory, and others exhibit rapid turnover or fluctuate during the cell cycle. Histone H4 Lys20 monomethylation (H4K20me1) has been shown to be involved in chromosome condensation, segregation, replication and repair. H4K20 methylation is controlled through a few methyltransferases, PR-Set7/Set8, SUV420H1, and SUV420H2, and a demethylase, PHF8. In cycling cells, the level of H4K20me1 increases during G2 and M phases and decreases during G1 phase. To monitor the local concentration and global fluctuation of histone modifications in living cells, we have developed a genetically encoded probe termed mintbody (modification-specific intracellular antibody; Sato et al., 2013 and 2016). By measuring the nuclear to cytoplasmic intensity ratio, the relative level of H4K20me1 in individual cells can be monitored. This detailed protocol allows the semi-quantitative analysis of the effects of methyltransferases on H4K20me1 levels in living cells based on H4K20me1-mintbody described by Sato et al. (2016).
Keywords: Post translational modification (翻译后修饰)Background
Posttranslational modifications of histone proteins play important roles in transcriptional regulation and genome integrity. While the one-dimensional epigenomic landscape has been revealed in many cell types by chromatin immunoprecipitation and sequencing, less is known about the dynamics of histone modifications due to technical limitations (Kimura et al., 2015). Recently, a few techniques for detecting protein modifications in living cells have been developed. One strategy uses sensors based on fluorescence/Förster resonance energy transfer (FRET) to monitor the balance between the modifying and demodifying enzymes. However, the dynamics of endogenous modifications cannot be monitored using FRET sensors. Another strategy that we have developed uses probes based on modification-specific antibodies. Fab-based live endogenous modification labeling (FabLEM) is a live-imaging system using fluorescently labeled antigen-binding fragments (Fabs). Fabs loaded into cells bind to the target modification without disturbing cell function as the binding time is very small (a second to tens of seconds). A genetically encoded system to express a modification-specific intracellular antibody (mintbody) can be applied for observation with a longer period of time or in living animals (Figure 1). Both Fabs and mintbodies are just small enough to pass through the nuclear pore by diffusion. When the level of the target modification increases, more probes become enriched in the nucleus. Therefore, by measuring the nuclear/cytoplasmic intensity ratio, changes of modification level in living cells can be monitored (Hayashi-Takanaka et al., 2011; Sato et al., 2013 and 2016). The live cell modification monitoring system using mintbodies will be particularly useful to evaluate the effects of small chemicals and protein depletion and overexpression.
Histone H4 Lys20 monomethylation (H4K20me1) is an essential modification in mammals, involved in chromosome condensation, segregation, replication and repair, as well as gene regulation (Beck et al., 2012; Jørgensen et al., 2013). The level of H4K20me1 increases during G2 to M phases and the inhibition of PR-Set7/Set8, a methyltransferase responsible for H4K20 monomethylation, causes mitotic defects. In female cells, the enrichment of H4K20me1 in inactive X chromosomes is microscopically observed. H4K20me1-specific mintbody has proven useful for monitoring the dynamic behavior of H4K20me1 in living cells (Sato et al., 2016). In addition, alteration of H4K20me1 level by ectopic expression of a methyltransferase has been evaluated. Among methyltransferases (PR-Set7/Set8, SUV420H1, and SUV420H2) and a demethylase (PHF8), involved in H4K20me1 metabolism, the expression of SUV420H1, which add methyl-groups to monomethylated H4K20 towards to trimethylation, caused a drastic effect. As an example of measuring relative H4K20me1 levels, we here describe the method to evaluate the effect of SUV420H1 on H4K20me1 in living cells.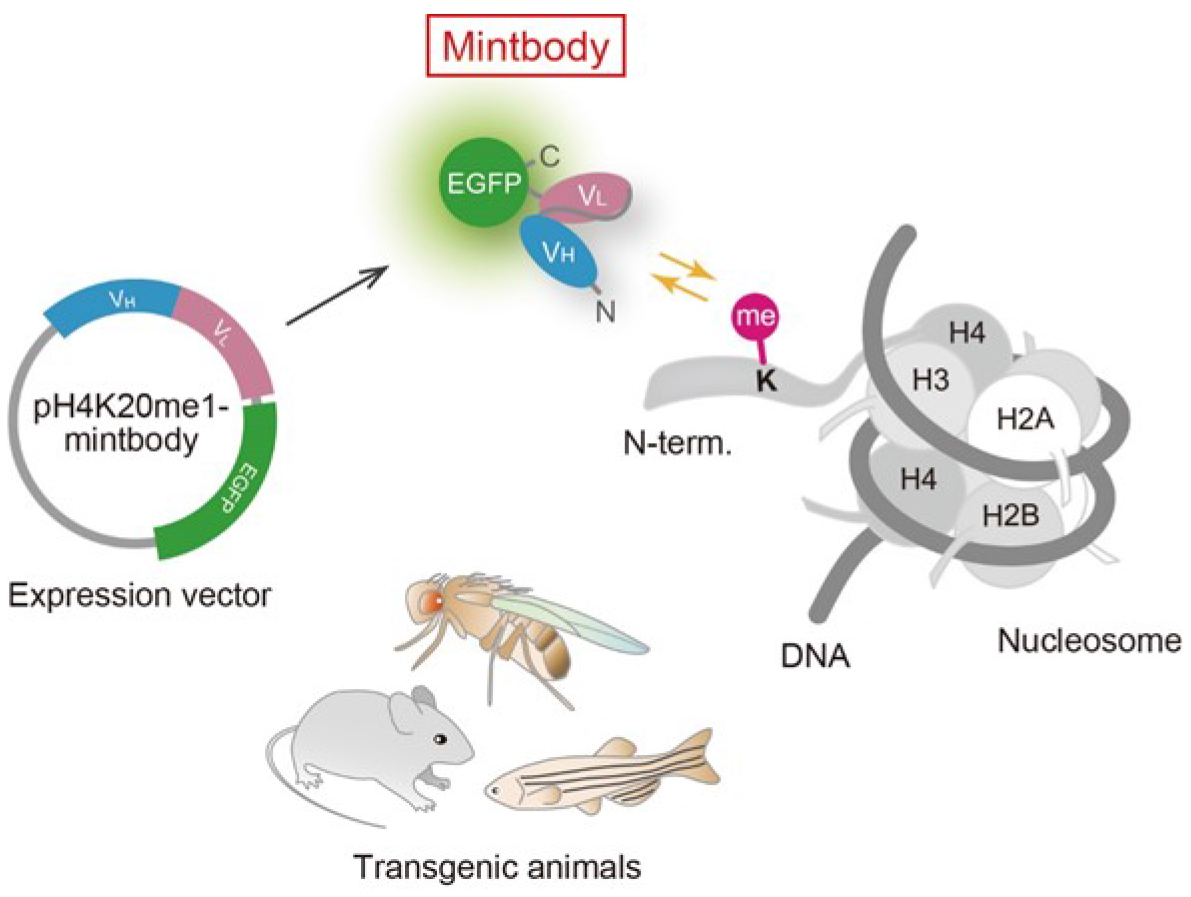
Figure 1. Schematic diagram of mintbody expression and function. A genetically encoded mintbody, which reversibly binds to specific modification, can be expressed in cells and animals that harbors the expression vector.
Materials and Reagents
- Pipette tips (10, 20, 200, 1,000 μl)
- 6 well plate (Corning, catalog number: 3516 )
- 10 cm dish (Greiner Bio One International, catalog number: 664160-013 )
- 24 well glass-bottom plate (IWAKI, catalog number: 5826-024 )
- HeLa cells (ATCC, catalog number: CRM-CCL-2 )
- Purified plasmid DNA (~1 μg/μl) encoding H4K20me1-mintbody based on pEGFP (Clontech) or a piggybac system (Sato et al., 2016) and Halo-SUV420H1 (Kazusa DNA Research Institute; FlexiHaloTag clone FHC01413)
- Dulbecco’s modified Eagle’s medium (DMEM), high glucose (4 g/L), containing L-Gln and sodium pyruvate (Nacalai Tesque, catalog number: 08458-16 )
- L-glutamine-penicillin-streptomycin solution (Sigma-Aldrich, catalog number: G1146-100ML )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- FuGENE HD (Promega, catalog number: E2312 )
- Opti-MEM media (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070 )
- G-418 disulfate aqueous solution (Nacalai Tesque, catalog number: 16513-26 )
- FluoroBrite DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: A1896701 )
- HaloTag TMR ligand (Promega, catalog number: G8251 )
Notes:
- Nucleotide sequence of H4K20me1-scFv is available in public databases (DDBJ/EMBL/GenBank) under the accession number LC129890. The plasmid DNA is available upon request to the authors.
- The original H4K20me1-specific antibody that was used to generate H4K20me1-mintbody (Hayashi-Takanaka et al., 2015) is available commercially (MBL International, catalog number: MABI0421).
Equipment
- CO2 incubator
- Pipette (10, 20, 200, 1,000 μl)
- 35 mm glass-bottom dishes, No. 1.5 coverslip (MATTEK, catalog number: P35G-1.5-14-C )
- Fluorescence microscope (Nikon Instruments, model: ECLIPSE Ti-E ) operated by NIS-elements and equipped with:
A spinning disk confocal unit (Yokogawa Electric, model: CSU-W1 )
An EM-CCD camera (Andor, model: iXon3 DU888 X-8465 )
An objective lens (Plan Apo 40x DIC M N2 [NA 0.95])
A laser unit (Nikon Instruments, model: LU-N4 )
A heated stage (Tokai Hit)
A CO2-control system (Tokken)
Software
- NIS-elements ver. 4.30 (Nikon Instruments)
- Excel (Microsoft)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sato, Y. and Kimura, H. (2017). Semi-quantitative Analysis of H4K20me1 Levels in Living Cells Using Mintbody. Bio-protocol 7(10): e2276. DOI: 10.21769/BioProtoc.2276.
分类
癌症生物学 > 通用技术 > 细胞生物学试验
细胞生物学 > 细胞染色 > 蛋白质
生物化学 > 蛋白质 > 修饰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











