- EN - English
- CN - 中文
Robust Generation of Knock-in Cell Lines Using CRISPR-Cas9 and rAAV-assisted Repair Template Delivery
使用CRISPR-Cas9和rAAV辅助的修复模板递送稳定生成敲入细胞系
发布: 2017年04月05日第7卷第7期 DOI: 10.21769/BioProtoc.2211 浏览次数: 23157
评审: Longping Victor TseRajesh B. ThippeshappaAnonymous reviewer(s)

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1216 阅读
Abstract
The programmable Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated nuclease 9 (Cas9) technology revolutionized genome editing by providing an efficient way to cut the genome at a desired location (Ledford, 2015). In mammalian cells, DNA lesions trigger the error-prone non-homologous end joining (NHEJ) DNA repair mechanism. However, in presence of a DNA repair template, Homology-Directed Repair (HDR) can occur leading to precise repair of the lesion site. This last process can be exploited to enable precise knock-in changes by introducing the desired genomic alteration on the repair template. In this protocol we describe the delivery of long repair templates (> 200 nucleotides) using recombinant Adeno Associated Virus (rAAV) for CRISPR-Cas9-based knock-in of a C-terminal tag sequence in a human cell line.
Keywords: CRISPR-Cas9 (CRISPR-Cas9)Background
Despite numerous reports on knock-out model systems generated by CRISPR-Cas9, knock-in reports are still lagging behind. Because of the many applications, generating knock-in cell lines remains an obvious goal of genome editing. The introduction of knock-in alterations generally relies on the presence of a repair template DNA and activation of the HDR repair mechanism after a site-specific double strand (ds)DNA break is introduced in the genome close to the site of alteration. Different templates can be delivered to the repair machinery ranging from a classical linearized vector containing extensive homology regions and an optional selection cassette, to single strand (ss)DNA oligonucleotides of about 200 nucleotides (Chen et al., 2011). Although ssDNA oligonucleotides are a popular tool, they can only be used to introduce small alterations such as mutations or epitope tags because of DNA synthesis limitations. In addition, the lack of a selection cassette requires robust screening strategies to identify correct clones as no selective pressure is applied on the HDR process. Successful use of integration-deficient rAAV for homologous recombination was already shown before the availability of tailored nucleases (Khan et al., 2011). Both its efficient delivery and ssDNA genome make rAAV a powerful tool to deliver donor repair templates for homologous recombination. Moreover, the secondary structures at the end of the ssDNA molecule block exonuclease activity and stabilize the donor DNA. Even without the use of specific nucleases, knock-in efficiencies of up to 0.7% were obtained in fibroblasts cells (Russell and Hirata, 1998), which was further increased by introducing selection cassettes.
By combining CRISPR-Cas9 with rAAV-mediated repair template delivery, knock-in cell lines can be generated in a robust manner with efficiencies well beyond 50% when selection cassettes are used. This protocol describes the complete procedure for epitope tagging of a gene of choice in the HCT116 colon carcinoma cell line using CRISPR-Cas9 and rAAV. A timeline for the complete experimental procedure is shown in Figure 1.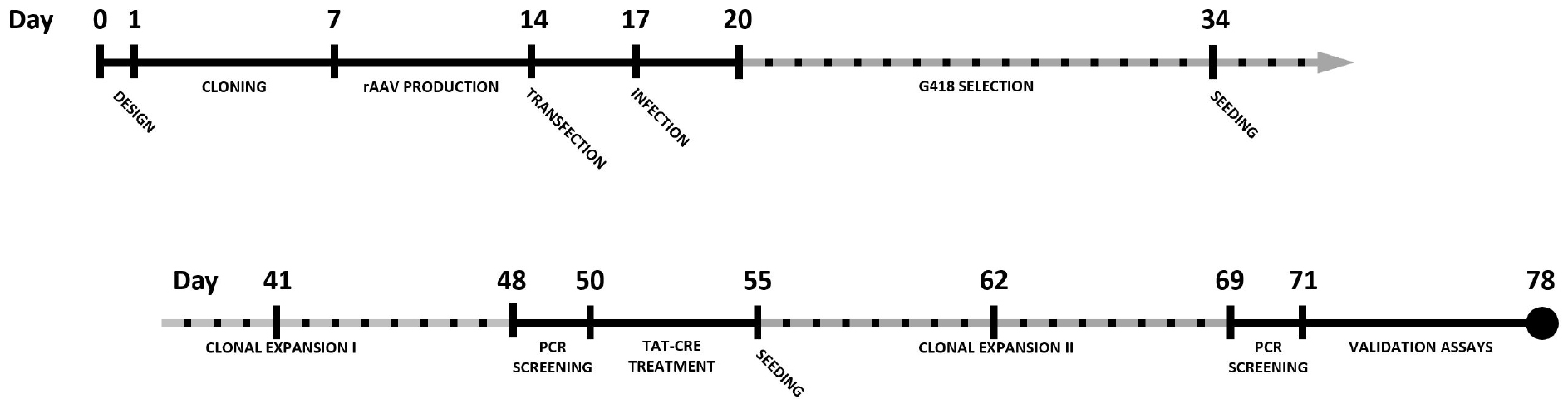
Figure 1. Timeline for the generation of a knock-in cell line using CRISPR-Cas9 and rAAV-assisted repair template delivery. Dotted timespans indicate periods of incubation or expansion, requiring limited to no hands-on time.
Materials and Reagents
- T25 cell culture treated flasks with filter caps (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 136196 )
- Eppendorf Safe-Lock microcentrifuge tubes (Eppendorf, catalog number: 022363204 )
- 5Prime Phase Lock gel tubes heavy 2 ml (Quantabio, catalog number: 2302830 )
- T75 cell culture treated flasks with filter caps (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 178905 )
- Greiner 50 ml centrifuge tubes (Greiner Bio One International, catalog number: 227261 )
- Greiner cell scrapers (Greiner Bio One International, catalog number: 541081 )
- 24-well cell culture-treated multidish (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 142475 )
- 96-well microplates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 156545 )
- 96-well PCR plate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: AB0700 )
- 6-well cell culture-treated multidish (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 140675 )
- Aluminium foil tape (3M, catalog number: 425DWB )
- Illustra Microspin S-400 HR columns (GE Healthcare, catalog number: 27-5140-01 )
- Immobilon-FL PVDF transfer membrane (EMD Millipore, catalog number: IPFL00010 )
- Whatman 3 MM chr cellulose blotting sheets (GE Healthcare, catalog number: 3030-917 )
- 0.22 µm filter (EMD Millipore, catalog number: SLGV033RS )
- AAV-293 cell line (Agilent Technologies, catalog number: 240073 )
- HCT 116 cell line (ATCC, catalog number: CCL-247 )
- Cas9 D10A nickase mutant (nCas9) (Addgene, catalog numbers: 48140 and 62987 )
- pAav-MCS-PQS1-3xFLAG or pAav-MCS-PQS2-3xHA (Addgene, catalog numbers: 84883 and 84917 respectively)
- pDG rAAV packaging plasmid (PlasmidFactory, catalog number: PF421 )
- Wild-type Cas9 expression vectors (Cas9) (Addgene, catalog numbers: 48138 and 62988 )
- Competent bacterial cells of choice (e.g., TOP10 or DH5α)
- Surveyor Mutation Detection Kit (Integrated DNA Technologies, catalog number: 706025 )
- UltraPure phenol:chloroform:isoamyl alcohol (25:24:1, v/v) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15593031 )
- Sodium chloride (NaCl) molecular biology grade (EMD Millipore, catalog number: 567441 )
- 2-propanol (Sigma-Aldrich, catalog number: 278475 )
- Ethanol (EMD Millipore, catalog number: 100983 )
- AccuPrime Pfx DNA polymerase (Thermo Fisher Scientific, InvitrogenTM, catalog number: 12344-024 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: 449709 )
- DMEM, high glucose GlutaMAX medium (Thermo Fisher Scientific, GibcoTM, catalog number: 31966047 )
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Benzonase nuclease (Sigma-Aldrich, catalog number: E1014 )
- AAV-purification Vira Kit 3-use (Virapur, catalog number: 003063 )
- AAV helper-free system (Agilent Technologies, catalog number: 240071 )
- McCoy’s 5A medium, modified (Thermo Fisher Scientific, GibcoTM, catalog number: 16600082 )
- Opti-MEM I reduced serum medium, GlutaMAX supplement (Thermo Fisher Scientific, GibcoTM, catalog number: 51985026 )
- Fugene HD transfection reagent (Promega, catalog number: E2311 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10500064 )
- Trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25300096 )
- Puromycin (Sigma-Aldrich, catalog number: P8833 )
- Crystal violet solution (Sigma-Aldrich, catalog number: HT90132 )
- Geneticin/G418 (Thermo Fisher Scientific, GibcoTM, catalog number: 11811031 )
- GoTaq G2 Hot Start polymerase (Promega, catalog number: M7405 )
- dNTP 100 mM PCR grade (Agilent Technologies, catalog number: 200415 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650 )
- TAT-Cre recombinase (Excellgen, catalog number: EG-1001 )
- NucleoSpin Gel and PCR Clean-up Kit (Machery-Nagel, catalog number: 740609 )
- Random Primer DNA Labeling Kit (Takara Bio, catalog number: 6045 )
- [α-32P] dCTP, 50 µCi (PerkinElmer, catalog number: BLU013H250UC )
- Exo-free Klenow enzyme (New England Biolabs, catalog number: M0212S )
- SuRE/Cut buffer H (Roche Diagnostics, catalog number: 11417991001 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A4503 )
- EcoRI 40 U/µl (Roche Diagnostics, catalog number: 10200310001 )
- Nytran SuPerCharge TurboBlotter Kit (Sigma-Aldrich, catalog number: Z613924 )
Note: This product has been discontinued. - PerfectHyb plus hybridization buffer (Sigma-Aldrich, catalog number: H7033 )
- 4-12% criterion XT Bis-Tris protein gel 18 well, 30 µl (Bio-Rad Laboratories, catalog number: 3450124 )
- Mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich, catalog numbers: F3165 )
Or rat monoclonal anti-HA high affinity (Roche Diagnostics, catalog number: 11867423001 ) - Zero Blunt PCR Cloning Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: K270020 )
- NucleoSpin Plasmid EasyPure (Machery-Nagel, catalog number: 740727 )
- Tris ultra-pure grade (MP Biomedicals, catalog number: 02103133 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: 436143 )
- Proteinase K from Tritirachium album > 800 U/ml (Sigma-Aldrich, catalog number: P4850 )
- HEPES (Sigma-Aldrich, catalog number: H4034 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: 255793 )
- Direct lysis reagent (cell) (VIAGEN BIOTECH, catalog number: 301-C )
- CHAPS hydrate (Sigma-Aldrich, catalog number: C5070 )
- cOmplete protease inhibitor (Roche Diagnostics, catalog number: 11697498001 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 30721 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 221465 )
- Sodium citrate tribasic dehydrate [HOC(COONa)(CH2COONa)2·2H2O] (Sigma-Aldrich, catalog number: C8532 )
- SmartLadder (Eurogentec, catalog number: MW-1700-10 )
- Phase Lock lysis buffer (see Recipes)
- TE-buffer (see Recipes)
- 2x HEBS (HEPES-buffered saline) (see Recipes)
- rAAV cell lysis buffer (see Recipes)
- Direct PCR lysis buffer (see Recipes)
- CHAPS lysis buffer (see Recipes)
- Depurination buffer (see Recipes)
- Denaturation buffer (see Recipes)
- Neutralization buffer (see Recipes)
- 20x SSC (see Recipes)
- Wash buffer 1 (see Recipes)
- Wash buffer 2 (see Recipes)
- Wash buffer 3 (see Recipes)
Equipment
- BSL2 cell culture facilities
- Thermoshaker
- Vortex (e.g., IKA, model: MS2 minishaker )
- Inverted microscope for cell culture use (e.g., Carl Zeiss, model: Axiovert 25 )
- Non-circulating water bath
- 250 ml sterile storage bottle (Corning, catalog number: 430281 )
- Thermal cycler instrument (e.g., Bio-Rad Laboratories, model: T100TM Thermal Cycler , catalog number: 1861096)
- Western blotting equipment, e.g.,
Criterion vertical electrophoresis cell (Bio-Rad Laboratories, catalog number: 1656001 )
Criterion blotter with plate electrodes (Bio-Rad Laboratories, catalog number: 1704070 ) - GS gene linker UV chamber (Bio-Rad Laboratories, model: GS Gene LinkerTM UV Chamber )
- Phosphor imager device (e.g., GE Healthcare, model: Typhoon 9200 )
- Required facilities and procedures (e.g., waste disposal procedures) should be in place for using radioactively labeled nucleotides ([α-32P] dCTP)
- Tabletop centrifuge (Eppendorf, model: 5430 R )
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Vandemoortele, G., De Sutter, D. and Eyckerman, S. (2017). Robust Generation of Knock-in Cell Lines Using CRISPR-Cas9 and rAAV-assisted Repair Template Delivery. Bio-protocol 7(7): e2211. DOI: 10.21769/BioProtoc.2211.
分类
分子生物学 > DNA > 诱/突变
分子生物学 > DNA > DNA 重组
细胞生物学 > 细胞工程 > CRISPR-cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link