- EN - English
- CN - 中文
Differential Salt Fractionation of Nuclei to Analyze Chromatin-associated Proteins from Cultured Mammalian Cells
从培养的哺乳动物细胞中差异盐分馏核分析染色质相关蛋白
发布: 2017年03月20日第7卷第6期 DOI: 10.21769/BioProtoc.2175 浏览次数: 17045
评审: Andrea PuharTatsuki KunohDavid A. Cisneros
Abstract
Nucleosomes are the core units of cellular chromatin and are comprised of 147 base pairs (bp) of DNA wrapped around an octamer of histone proteins. Proteins such as chromatin remodelers, transcription factors, and DNA repair proteins interact dynamically with chromatin to regulate access to DNA, control gene transcription, and maintain genome integrity. The extent of association with chromatin changes rapidly in response to stresses, such as immune activation, oxidative stress, or viral infection, resulting in downstream effects on chromatin conformation and transcription of target genes. To elucidate changes in the composition of proteins associated with chromatin under different conditions, we adapted existing protocols to isolate nuclei and fractionate cellular chromatin using a gradient of salt concentrations. The presence of specific proteins in different salt fractions can be assessed by Western blotting or mass spectrometry, providing insight into the degree to which they are associated with chromatin.
Keywords: Chromatin (染色质)Background
Many chromatin-associated proteins are insoluble under low salt conditions because of their charged-based interaction with DNA or histones. Since salt disrupts charged-based protein-DNA and protein-protein interactions, chromatin-associated proteins become more soluble with increasing concentration of NaCl (Teves and Henikoff, 2012). Proteins strongly bound to DNA are expected to elute with high salt whereas loosely bound proteins, such as transcription factors, will elute with low salt. We are specifically interested in how virus infection alters the composition of factors associated with the cellular chromatin. Nuclear replicating viruses, such as adenovirus, herpes simplex virus, and Epstein-Barr virus, dramatically alter the appearance of the host chromatin during infection (Avgousti et al., 2016; Lam et al., 2010; Simpson-Holley et al., 2005; Chiu et al., 2013). We hypothesized that these changes in appearance are partly due to differences in protein composition of host chromatin. Changes in host chromatin could reflect antiviral defenses mounted by the cell or active manipulation by the virus. To compare association of proteins with chromatin in uninfected and infected cells we developed this protocol to fractionate nuclei using a salt gradient (Figure 1). In this protocol we isolate nuclei, digest the DNA down to mono-nucleosome length, and then wash the nuclei with increasing concentrations of salt, collecting each fraction for analysis by Western blotting. We recently used this protocol to elucidate changes to cellular chromatin during infection with adenovirus (Avgousti et al., 2016). We now present this protocol as a general approach to monitor association of proteins with chromatin under a wide range of perturbing conditions. 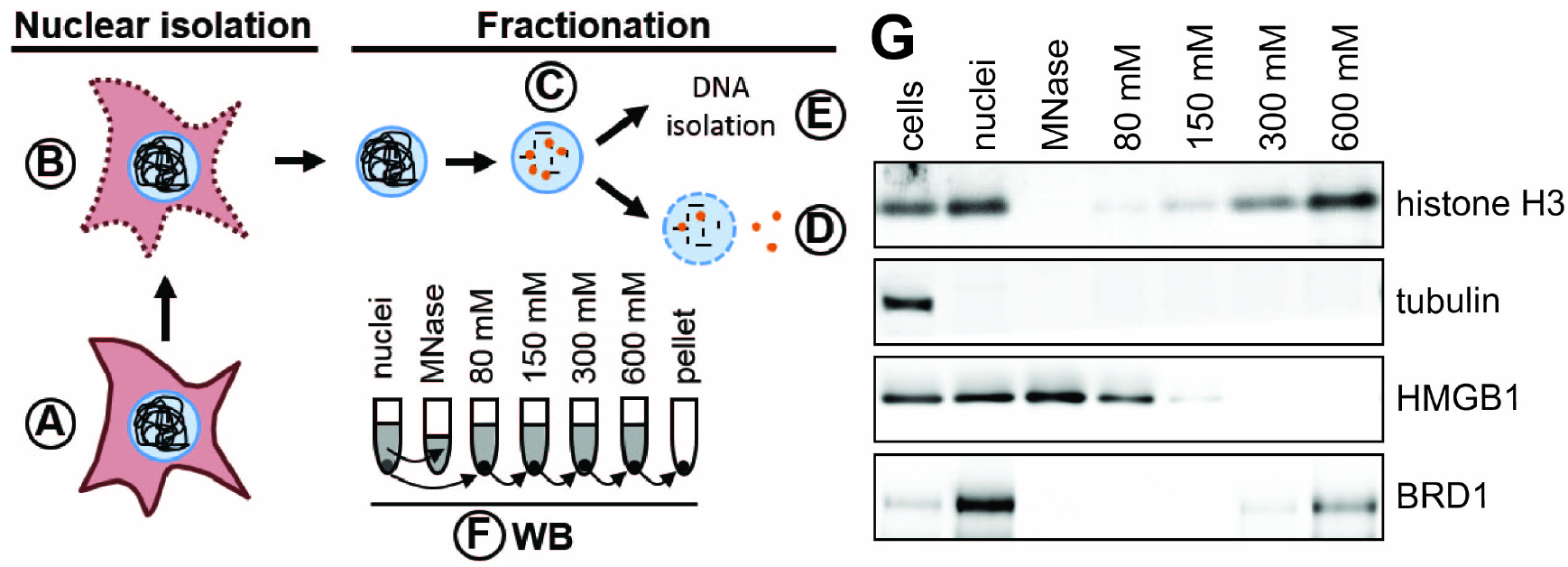
Figure 1. Schematic of nuclear fractionation and example Western blot. A. Roughly 4 x 107 cells are prepared per condition. B. Plasma membranes are permeabilized and nuclei are isolated either by sucrose cushion (step B1) or using a Dounce homogenizer (step B2). C. DNA is digested to mono-nucleosome length using MNase. Proteins loosely bound to chromatin elute during this step. D. The chromatin is further fractionated by washing the nuclei in buffers with increasing salt concentration. E. The DNA is isolated from nuclei to confirm digestion of the cellular genome to 150 bp fragments. F. The quality of fractionation is tested using SDS-PAGE and Western blot (WB) for control proteins (e.g., tubulin, histone H3). The grey colored supernatants (and the pellet in case of the nuclei) represent the samples used for Western blot analysis. G. Example Western blot analysis of chromatin fractionation. Tubulin is found only in the cytoplasmic fraction and is a suitable control to test the quality of nuclear isolation. Histone H3 is a component of cellular chromatin and only elutes from nuclei in buffers with high salt. HMGB1 is a highly mobile nuclear protein (Sapojnikova et al., 2005) and thus elutes during MNase digest and under lower salt conditions. Brd1 directly binds to histone tails (Sanchez et al., 2014) and elutes under high salt conditions.
Materials and Reagents
Note: Comparable reagents from different suppliers may be used for the protocol.
- 150 mm tissue culture dishes (Corning, Falcon®, catalog number: 353025 )
- 15 ml centrifuge tube (Corning, catalog number: 430790 )
- 5 ml pipettes (VWR, catalog number: 89130-908 )
- Transfer pipette (Denville Scientific, catalog number: P7222 )
- 30 ml glass tube (Corning, Corex®, catalog number: 1-8445-30 )
Note: This product has been discontinued. - 1.7 ml microcentrifuge tubes (VWR, catalog number: 87003-294 )
- Pipette tips
0.1-10 µl (Corning, catalog number: 4153 )
1-200 µl (Corning, catalog number: 4126 )
100-1,000 µl (Corning, catalog number: 4129 ) - 250 ml sterile disposable filter units with 0.2 µm PES membrane (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 568-0020 ) (used for Buffer I and Buffer II)
- 60 ml syringe (BD, catalog number: 309653 ) (used for Buffer IV.80, IV.150, IV.300 and IV.600)
- 25 mm syringe filter (Pall, catalog number: 4612 ) (used for Buffer IV.80, IV.150, IV.300 and IV.600)
- A549 cells (ATCC, catalog number: CCL-185 )
- Ham’s F-12K cell culture media (Thermo Fisher Scientific, GibcoTM, catalog number: 21127-022 )
- Fetal bovine serum (FBS) (VWR, catalog number: 89510-182 )
- Penicillin-streptomycin (Pen/Strep) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140-122 )
- Trypsin-EDTA (0.25%) (Thermo Fisher Scientific, GibcoTM, catalog number: 25200-056 )
- Phosphate buffered saline (PBS) (Mediatech, catalog number: 21-030-CM )
- Liquid nitrogen
- NP-40/IGEPAL® CA-630 (Sigma-Aldrich, catalog number: I8896 ) (10% stock solution in H2O)
- Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 ) (0.1 M stock solution in isopropanol)
- 1,4-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 10197777001 ) (1 M stock solution in HEPES buffer, pH 7.75)
- Protease inhibitor cocktail (Roche Diagnostics, catalog number: 11697498001 ) (prepared as 50x stock solution in H2O according to manufacturer instructions)
- Micrococcal nuclease (MNase) (Sigma-Aldrich, catalog number: N3755 ) (0.2 U/µl stock solution in H2O)
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 ) (0.1 mM stock solution in H2O, pH = 10)
- PCR purification kit (QIAGEN, catalog number: 28104 )
- Orange G (Sigma-Aldrich, catalog number: O3756 ) (0.35% [w/v] orange G with 30% [w/v] sucrose in H2O for 6x stock solution)
- 100 bp DNA ladder (New England Biolabs, catalog number: N3231 )
- Broad range protein ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 26623 )
- GelRed nucleic acid gel stain (Biotum, catalog number: 41003 )
- LDS sample buffer (4x) (Thermo Fisher Scientific, NovexTM, catalog number: NP0007 )
- Sucrose (Fisher Scientific, catalog number: BP220-1 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 ) (1 M stock solution in H2O)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9625 ) (5 M stock solution in H2O)
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670 ) (1 M stock solution in H2O)
- Trizma base (Sigma-Aldrich, catalog number: T1503 ) (1 M stock solution in H2O adjusted to pH 7.4 with HCl)
- UltraPure agarose (Thermo Fisher Scientific, InvitrogenTM, catalog number: 16500500 )
- Hydrochloric acid 6.0 N solution (HCl) (Fisher Scientific, catalog number: MK-H168-4 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C5080 ) (0.5 M stock solution in H2O)
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- HEPES (Sigma-Aldrich, catalog number: H3375 ) (1 M stock solution in H2O adjusted to pH 7.9 with NaOH)
- Sodium hydroxide (NaOH) (AMRESCO, catalog number: M137 )
- Buffer I.A and I.B (see Recipes)
- Buffer II (see Recipes)
- Buffer III.A and III.B (see Recipes)
- Buffer IV.80, IV.150, IV.300 and IV.600 (see Recipes)
- Hypotonic buffer (see Recipes)
Equipment
Note: Equipment with similar properties may be used for the protocol, however, we recommend using a specific kind of reusable centrifuge tubes (listed in 5) to ensure high quality isolation of nuclei.
- CO2 incubator for cell culture (BINDER, catalog number: 9040-0082 )
- Benchtop centrifuge (Beckman Coulter, model: Allegra X-14R )
- Rotors for benchtop centrifuge (Beckman Coulter, models: SX4750 for tissue culture and FX6100 for 10,000 x g spins, or seminal rotors suitable for high speeds)
- Adapters for FX6100 rotor (Beckman Coulter, catalog number: 392830 )
- 30 ml reusable centrifuge tubes (Sigma-Aldrich, catalog number: T2793 )
- Tabletop centrifuge 5424 R (Eppendorf, model: 5424 R )
- 1 ml tissue grinder (Dounce homogenizer) with tight fitting pestle (Ace Glass Incorporated, catalog number: 8343-01 )
- Water bath (Fisher Scientific, model: IsotempTM Digital-Control Water Baths Model 215 , catalog number: 15-462-15Q)
- Tube rotator (VWR, catalog number: 10136-084 )
- Pipettes
1-10 µl (Gilson, catalog number: F144055P )
2-20 µl (Gilson, catalog number: F144056M )
20-200 µl (Gilson, catalog number: F144058M )
100-1,000 µl (Gilson, catalog number: F144059M ) - Agarose gel electrophoresis systems (Thermo Fisher Scientific, Thermo ScientificTM, model: Owl EasyCast B1A system )
- Fluorescence and chemiluminescence gel imaging system (Syngene, model: G: BOX Chemi XT4 )
- Heat block (Fisher Scientific, model: IsotempTM Digital Dry Baths/Block Heaters , catalog number: 88-860-022)
- Protein electrophoresis apparatus (Bio-Rad Laboratories, model: Mini-PROTEAN® Tetra Vertical Electrophoresis Cell for Mini Precast Gels , catalog number: 1658005)
- Western blot apparatus (Thermo Fisher Scientific, model: SureLockTM Mini-Cell Electrophoresis System )
Software
- ImageJ (freely available from National Institutes of Health, https://imagej.nih.gov/ij/)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Herrmann, C., Avgousti, D. C. and Weitzman, M. D. (2017). Differential Salt Fractionation of Nuclei to Analyze Chromatin-associated Proteins from Cultured Mammalian Cells. Bio-protocol 7(6): e2175. DOI: 10.21769/BioProtoc.2175.
分类
免疫学 > 宿主防御 > 综合
癌症生物学 > 癌症生物化学 > 蛋白质
细胞生物学 > 细胞器分离 > 细胞核
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













