- EN - English
- CN - 中文
Multiplexed GuideRNA-expression to Efficiently Mutagenize Multiple Loci in Arabidopsis by CRISPR-Cas9
多重向导RNA表达通过CRISPR Cas9对拟南芥中的多个基因座进行高效诱变
发布: 2017年03月05日第7卷第5期 DOI: 10.21769/BioProtoc.2166 浏览次数: 12967
评审: Rainer MelzerMarta BjornsonMoritz BomerDaniel Savatin
Abstract
Since the discovery of the CRISPR (clustered regularly interspaced short palindromic repeats)-associated protein (Cas) as an efficient tool for genome editing in plants (Li et al., 2013; Shan et al., 2013; Nekrasov et al., 2013), a large variety of applications, such as gene knock-out, knock-in or transcriptional regulation, has been published. So far, the generation of multiple mutants in plants involved tedious crossing or mutagenesis followed by time-consuming screening of huge populations and the use of the Cas9-system appeared a promising method to overcome these issues. We designed a binary vector that combines both the coding sequence of the codon optimized Streptococcus pyogenes Cas9 nuclease under the control of the Arabidopsis thaliana UBIQUITIN10 (UBQ10)-promoter and guide RNA (gRNA) expression cassettes driven by the A. thaliana U6-promoter for efficient multiplex editing in Arabidopsis (Yan et al., 2016). Here, we describe a step-by-step protocol to cost-efficiently generate the binary vector containing multiple gRNAs and the Cas9 nuclease based on classic cloning procedure.
Keywords: CRISPR-Cas9 (CRISPR-Cas9)Background
The RNA-guided Cas9-system is derived from the bacterial defense system against foreign DNA (Sorek et al., 2013). It has been recognized as a method of choice for genome editing because of its high efficiency, easy handling and possibility of multiplex editing. In general, the Cas9-gene editing system involves a single synthetic RNA molecule, the gRNA that directs the Cas9 protein to target the desired DNA site for genome modification or transcriptional control. The gRNA-Cas9 complex recognizes the targeted DNA by gRNA-DNA pairing and requires the presence of a protospacer-adjacent motif (PAM). The PAM is represented by the nucleotides NGG or less specific NAG (with N for any nucleotide) in the target site following the gRNA-DNA pairing region. Thus, the approximate 20 nucleotides long gRNA spacer sequence, i.e., the part of the gRNA sequence complementary to the DNA target site, determines the specificity of the complex. In this protocol, we describe the details to generate a binary vector that contains both the gRNA and the Cas9 coding sequence by classic cloning (Figure 1). As our vector system allows for subsequent addition of further gRNAs, it can be used for multiplex editing of the Arabidopsis genome and to obtain multiple, stably inherited alleles. Strong expression of the Cas9 protein and the gRNAs especially in proliferating tissues is achieved by the use of the A. thaliana UBQ10- and the U6-promoter, respectively. First mutations can be detected in the T1 generation, and T-DNA- and Cas9-free mutant plants may already be selected in the T2 generation. Besides the selection of the gRNA and the construction of the plasmid, we give an overview of efficient genotyping methods required for detection of small or large deletions.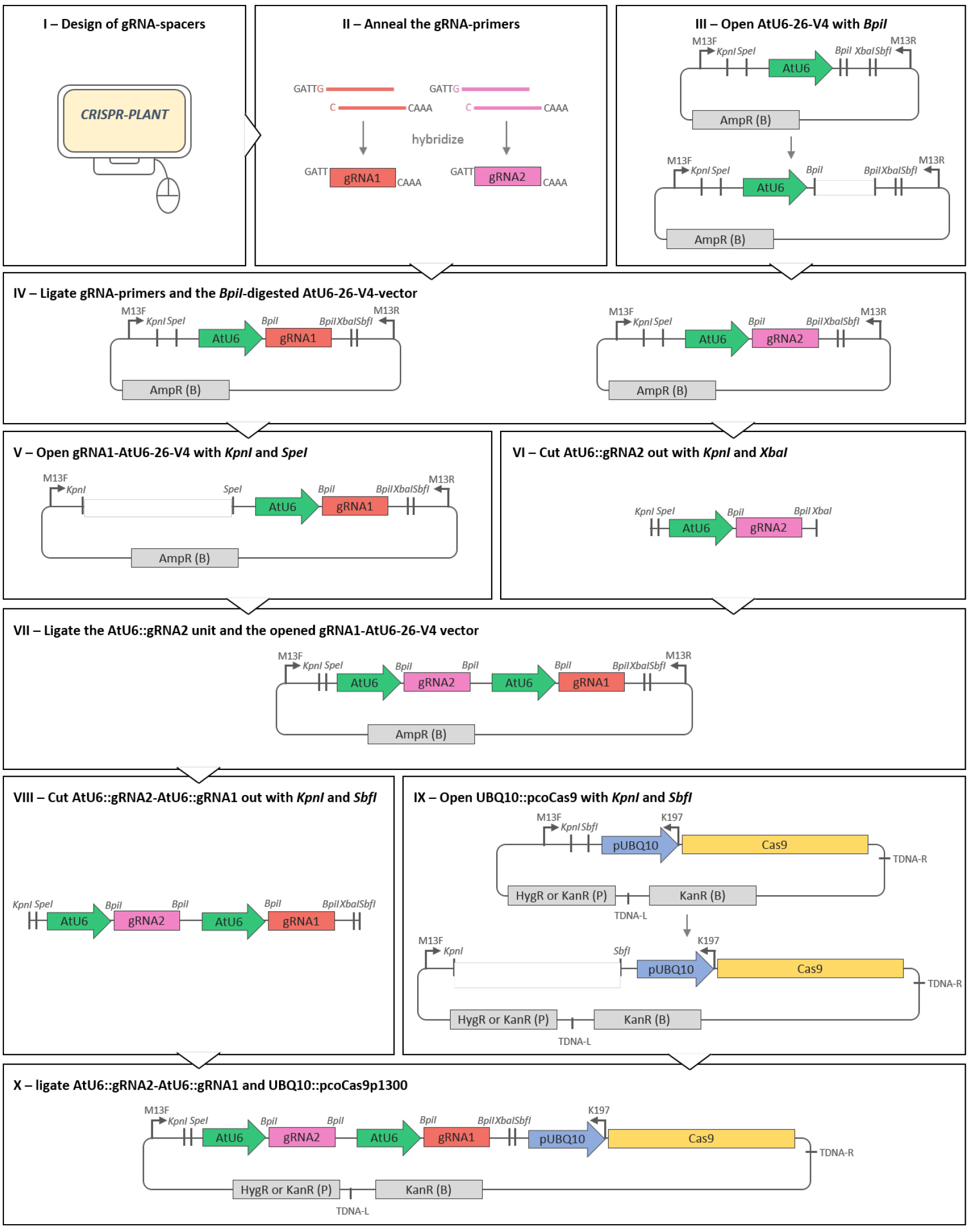
Figure 1. Scheme of the cloning procedure described in the protocol. Only a series of restriction and ligation steps is required to obtain a plant transformation vector equipped with a set of AtU6-driven gRNAs and the Cas9 enzyme under the control of the UBQ10 promoter. Restriction enzyme cutting sites are displayed in italics. Arrows on vectors indicate primer binding sites. TDNA-R and TDNA-L point out T-DNA right and left borders, respectively. (B) and (P) indicate selection markers for selection in bacteria or plants, respectively.
Materials and Reagents
Note: The protocol described here is based on the RNA-guided Cas9 system which was published recently (Yan et al., 2016). Only classical cloning methods such as restriction enzyme digestion and cohesive end ligation are required to construct plasmids ready for plant transformation.
- Consumables
- 200 µl PCR tubes (Kisker Biotech, catalog number: G003-SF )
or 96-well PCR plates (SARSTEDT, catalog number: 72.1978.202 )
StarSeal sealing tape (STARLAB, catalog number: E2796-9793 ) - 1.5 ml reaction tubes (SARSTEDT, catalog number: 72.690.001 )
- Pipette tips
- Scalpel
- Petri dishes, round, 9.2 x 1.6 cm (SARSTEDT, catalog number: 82.1472.001 )
- Petri dishes, square, 10 x 10 x 2 cm (SARSTEDT, catalog number: 82.9923.422 )
- Cuvettes for electroporation, e.g., Gene Pulser® cuvette 0.1 cm (Bio-Rad Laboratories, catalog number: 1652089 )
- 5 ml glass pipette
- MicroporeTM tape (VWR, catalog number: 115-8172 )
Note: Any appropriate consumable can be used. - Competent cells
- One Shot® TOP10 chemically competent Escherichia coli (Thermo Fisher Scientific, InvitrogenTM, catalog number: C4040 )
- Electro-competent Agrobacterium tumefaciens, strain pGV3101 (for preparation of electro-competent Agrobacteria, please see Mersereau et al., 1990)
- Plant material
- Arabidopsis thaliana Col-0
- Plasmids
- AtU6-26-V4 (3.5 kb, ampicillin resistance marker [AmpR], available on request)
- UBQ10::pcoCas9p1300 (14.3 kb, kanamycin resistance marker [KanR] in bacteria, hygromycin or kanamycin resistance marker [HygR or KanR] in plants, available on request)
- Optional: pGEM®-T easy (Promega, catalog number: A3600 )
- Enzymes and buffers
Restriction- BpiI (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: ER1011 )
- KpnI-HF (New England Biolabs, catalog number: R3142 )
- XbaI (New England Biolabs, catalog number: R0145 )
- SpeI-HF (New England Biolabs, catalog number: R3133 )
- SbfI-HF (New England Biolabs, catalog number: R3642 )
- Cutsmart® buffer (New England Biolabs, catalog number: B7204S , supplied with the enzyme)
Genotyping (bacteria) - BpiI (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: ER1011 )
- Green Taq DNA polymerase (GenScript, catalog number: E00043 )
- 10x Taq buffer (GenScript, catalog number: B0005 , supplied with the enzyme)
- 10 mM dNTPs (Carl Roth, catalog number: K039 )
- Any DNA-loading dye, e.g., 6x gel loading dye, purple (New England Biolabs, catalog number: B7024 )
- Any DNA-size standard, e.g., GeneRulerTM 1 kb Plus DNA ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM1331 )
- Agarose (Carl Roth, catalog number: 3810 )
- 10x TBE electrophoresis buffer (see Recipes)
Tris (Applichem, catalog number: A1379 )
Boric acid (Applichem, catalog number: 131015 )
EDTA (Applichem, catalog number: 131669 ) - Deionized water (sterile)
- Antibiotics (store stocks at -20 °C)
- Ampicillin (Carl Roth, catalog number: K029 , stock 50 mg/ml in deionized water)
- Kanamycin (Carl Roth, catalog number: T832 , stock 30 mg/ml in deionized water)
- Rifampicin (Applichem, catalog number: A2220 , stock 25 mg/ml in DMSO)
- Gentamycin (Carl Roth, catalog number: 0233 , stock 10 mg/ml in deionized water)
- Hygromycin (Carl Roth, catalog number: CP13 , stock 10 mg/ml in deionized water)
- Media (see Recipes)
- YEB medium for Agrobacterium
Meat extract (Carl Roth, catalog number: 5770 )
Yeast extract (Carl Roth, catalog number: 2904 )
Peptone (Sigma-Aldrich, catalog number: 82303 )
Sucrose (Carl Roth, catalog number: 4621 )
Magnesium sulfate (MgSO4) (Carl Roth, catalog number: P027 )
Bacto-agar (Th. Geyer, CHEMSOLUTE®, catalog number: 9914-500G )
Sodium hydroxide (NaOH) (Merck, catalog number: 28245 ) - YT medium for E. coli
Sodium chloride (NaCl) (Carl Roth, catalog number: 9265 )
Yeast extract (Carl Roth, catalog number: 2904)
Peptone (Sigma-Aldrich, catalog number: 82303)
Bacto-agar (Th. Geyer, CHEMSOLUTE®, catalog number: 9914-500G)
Sodium hydroxide (NaOH) (Merck, catalog number: 28245) - ½ MS for Arabidopsis
MS + B5 Vitamins (Duchefa Biochemie, catalog number: M0231 )
Potassium hydroxide (KOH) (Merck Millipore, catalog number: 105033 ) - DNA purification
- Gel-purification, e.g., NucleoSpin® Gel and PCR clean-up (MACHEREY-NAGEL, catalog number: 740609 )
- Plasmid isolation, e.g., NucleoSpin® Plasmid EasyPure (MACHEREY-NAGEL, catalog number: 740727 )
- Silwet L-77 (Lehle seeds, catalog number: VIS-30 ) for plant transformation
- Bleach (Carl Roth, catalog number: 9062 )
- 37% HCl (Merck Millipore, catalog number: 100317 )
- Genotyping (plants)
- Optional: Phire Plant Direct PCR Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: F-130WH )
- Optional: T7 endonuclease I (New England Biolabs, catalog number: M0302 ) plus NEBuffer 2 (New England Biolabs, catalog number: B7002 ), 250 mM EDTA
- Optional: pGEM®-T easy (Promega, catalog number: A3600)
- Oligonucleotides (5’-3’), 10 pmol/µl
- M13F_TGTAAAACGACGGCCAGT
- M13R_CAGGAAACAGCTATGACC
- K197 _CTGTTAATCAGAAAAACTCAG
Equipment
Note: No specific equipment is required. Any appropriate device can be used.
- Computer with internet access
- Thermocycler (e.g., Eppendorf, model: Mastercycler® nexus )
- Thermoblock (e.g., Eppendorf, model: Thermomixer® comfort )
- Plate incubators (28 °C, 37 °C)
- Shakers (28 °C, 37 °C)
- Horizontal gel-electrophoresis system (e.g., Bio-Rad Laboratories, model: Mini-Sub® Cell GT System )
- UV transilluminator (e.g., Bio-Rad Laboratories, model: GelDocTM XR+ System )
- Electroporator, (e.g., Bio-Rad Laboratories, model: MicroPulserTM Electroporator )
- 10 L desiccator
- Fume hood
- 200 ml beaker
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Schumacher, J., Kaufmann, K. and Yan, W. (2017). Multiplexed GuideRNA-expression to Efficiently Mutagenize Multiple Loci in Arabidopsis by CRISPR-Cas9. Bio-protocol 7(5): e2166. DOI: 10.21769/BioProtoc.2166.
分类
植物科学 > 植物转化 > 农杆菌介导的转化方法
植物科学 > 植物发育生物学 > 综合
分子生物学 > DNA > DNA 修饰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












