- EN - English
- CN - 中文
Expression and Purification of the GRAS Domain of Os-SCL7 from Rice for Structural Studies
表达、纯化水稻Os-SCL7的GRAS结构域用于结构研究
发布: 2017年02月05日第7卷第3期 DOI: 10.21769/BioProtoc.2122 浏览次数: 8513
评审: Arsalan DaudiAnonymous reviewer(s)

相关实验方案
利用SP3珠和稳定同位素质谱技术优化蛋白质合成速率:植物核糖体的案例研究
Dione Gentry-Torfer [...] Federico Martinez-Seidel
2024年05月05日 2843 阅读

基于活性蛋白质组学和二维聚丙烯酰胺凝胶电泳(2D-PAGE)鉴定拟南芥细胞间隙液中的靶蛋白酶
Sayaka Matsui and Yoshikatsu Matsubayashi
2025年03月05日 1949 阅读
Abstract
GRAS proteins, named after the first three members GAI, RGA and SRC, has been found in 294 embryophyta species and is represented by 1,035 sequences. They belong to a plant-specific protein family and play essential roles in plant growth and development. Proteins in this family are defined as minimally containing a conserved GRAS domain, which is about 350-450 resides and can be subdivided into five distinct motifs with their name derived from the most prominent amino acids: LRI (leucine-rich region I), VHIID, LRII (leucine-rich region II), PFYRE and SAW and mainly function in the interaction between GRAS proteins and their partners (Sun et al., 2012).By phylogenetic analysis, the GRAS family can be divided into more than ten subfamilies, of which SCL4/7 is one important subgroup and functions in response to environmental stresses. Here we describe a detailed protocol for the expression and purification of the GRAS domain of Os-SCL7, a SCL4/7 member in rice, which enables us to crystallize it and determine its structure.
Keywords: Expression (表达)Background
The GRAS proteins are a large family that plays vital roles in plant development and signaling transduction. Findings indicate that some family members such as DELLAs function as a repressor of GA responsive plant growth and are key regulatory targets in the GA signaling pathway (Murase et al., 2008), NSP1 and NSP2 play important roles in regulating nodulation development and signaling (Kaló et al., 2005), the proteins SCR and SHR together play an important role in the control of radial patterning for both the root and shoot (Helariutta et al., 2000), AtLAS is a key regulator in the developmental processes of the axillary meristem (Greb et al., 2003), HAM functions in shoot meristem maintenance (Stuurman et al., 2002).
Based on sequence analyses, GRAS proteins include a variable N-terminal domain and a widely and highly conserved C-terminal domain known as the GRAS domain. The N-terminal domains constitute a plant-specific unfoldome and may act as molecular bait by initiating the key molecular recognition events (Uversky et al., 2010). And the C-terminal GRAS domain is highly conserved in the whole GRAS family, suggesting that these proteins share a similar function and/or a common mode-of-action (Sun et al., 2012).
Though many members of GRAS proteins have been studied, the functional mechanism of GRAS proteins is still unclear. Structural descriptions of GRAS proteins may deeply clarify the functional mechanism of this family. However as yet little structural analyses have been reported, mainly due to the difficulties in obtaining sufficient quality and quantity of GRAS proteins. Soto et al. (2014) reported the expression and purification of the GRAS domain of rice SLR1. They constructed a GST-SLR1 fusion protein and expressed it in E. coli. But the expression levels were low (0.5 mg [TB medium] or 0.2 mg [M9 medium] of purified protein from 1 L flask culture). With some modifications, they obtained 1-3 mg of stable isotope labeled purified protein at 87% purity from 1 L of fermenter culture. However, the expression levels and purity of SLR1 are both not enough for crystallization. Moreover, reducing the protein synthesis rate by low culture temperature and speed, co-expressing with chaperonins, expressing as a fusion protein with soluble tags such as glutathione S-transferase, thioredoxin and maltose-binding protein or mutating some hydrophobic or disulfide forming amino acids are usually used to improve the expression and solubility of target proteins. While using more purification methods and steps will help to improve the purity of target proteins. In order to get a high purity and quantity of GRAS protein, we established a protocol for easy expression and purification of the GRAS domain of Os-SCL7 from rice.
Materials and Reagents
- Amicon Ultra centrifugal filters, 30 K (Merck Millipore, catalog number: UFC903096 )
- Amicon Ultra centrifugal filters 30,000 MWCO (EMD Millipore, catalog number: UFC803024 )
- Economical biotech dialysis membrane, 14 KD (Sangon Biotech, catalog number: TX0111 )
- Escherichia coli (E. coli) (BL21) (Thermo Fisher Scientific, InvitrogenTM, catalog number: C6000-03 )
- pET32aM (Novagen, modified from pET32a by inserting a TEV protease cut site inside the NcoI cleavage site) (Figure 1)
- Rice (NM_001057650, residues 201-578)
- Ampicillin (Sangon Biotech, catalog number: A100741 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sangon Biotech, catalog number: A100487 )
- Chelating Sepharose Fast FLow (GE Healthcare, catalog number: 17057502 )
- Nickel(II) sulfate hexahydrate (NiSO4·6H2O) (Sangon Biotech, catalog number: A600658 )
- Imidazole (Sangon Biotech, catalog number: A500529 )
- TEV (tobacco etch virus) protease (produced in-house) (Fang et al., 2007)
- Tryptone (Oxoid, catalog number: LP0042 )
- Yeast extract (Oxoid, catalog number: LP0021 )
- Sodium chloride (NaCl) (Sangon Biotech, catalog number: A501218 )
- Agar (Oxoid, catalog number: LP0011 )
- Tris-base (Sangon Biotech, catalog number: A100826 )
- Hydrochloric acid (HCl) (Sinopharm Chemical Reagent, catalog number: 7647-01-0 )
- Glycerol (Sangon Biotech, catalog number: A100854 )
- Ethylenediaminetetraacetic acid disodium salt (EDTA) (Sangon Biotech, catalog number: A100105 )
- β-mercaptoethanol (AMRESCO, catalog number: 60-24-2 )
- Tween-20 (Sangon Biotech, catalog number: A100777 )
- Ethanol anhydrous (Sangon Biotech, catalog number: A500737 )
- Phosphoric acid ortho (85%) (Sangon Biotech, catalog number: A502803 )
- Acetic acid (Sangon Biotech, catalog number: A501931 )
- Coomassie Brilliant Blue G-250 (Sangon Biotech, catalog number: A100615 )
- Coomassie Brilliant Blue R-250 (Sangon Biotech, catalog number: A100472 )
- SDS (Sangon Biotech, catalog number: A100227 )
- Isopropanol (Sangon Biotech, catalog number: A507048 )
- LB medium (see Recipes)
- LB-agar plates (see Recipes)
- Lysis buffer (see Recipes)
- Chromatography buffer (see Recipes)
- Coomassie Brilliant Blue G-250 buffer (see Recipes)
- SDS-PAGE staining solution (see Recipes)
- SDS-PAGE destaining solution (see Recipes)
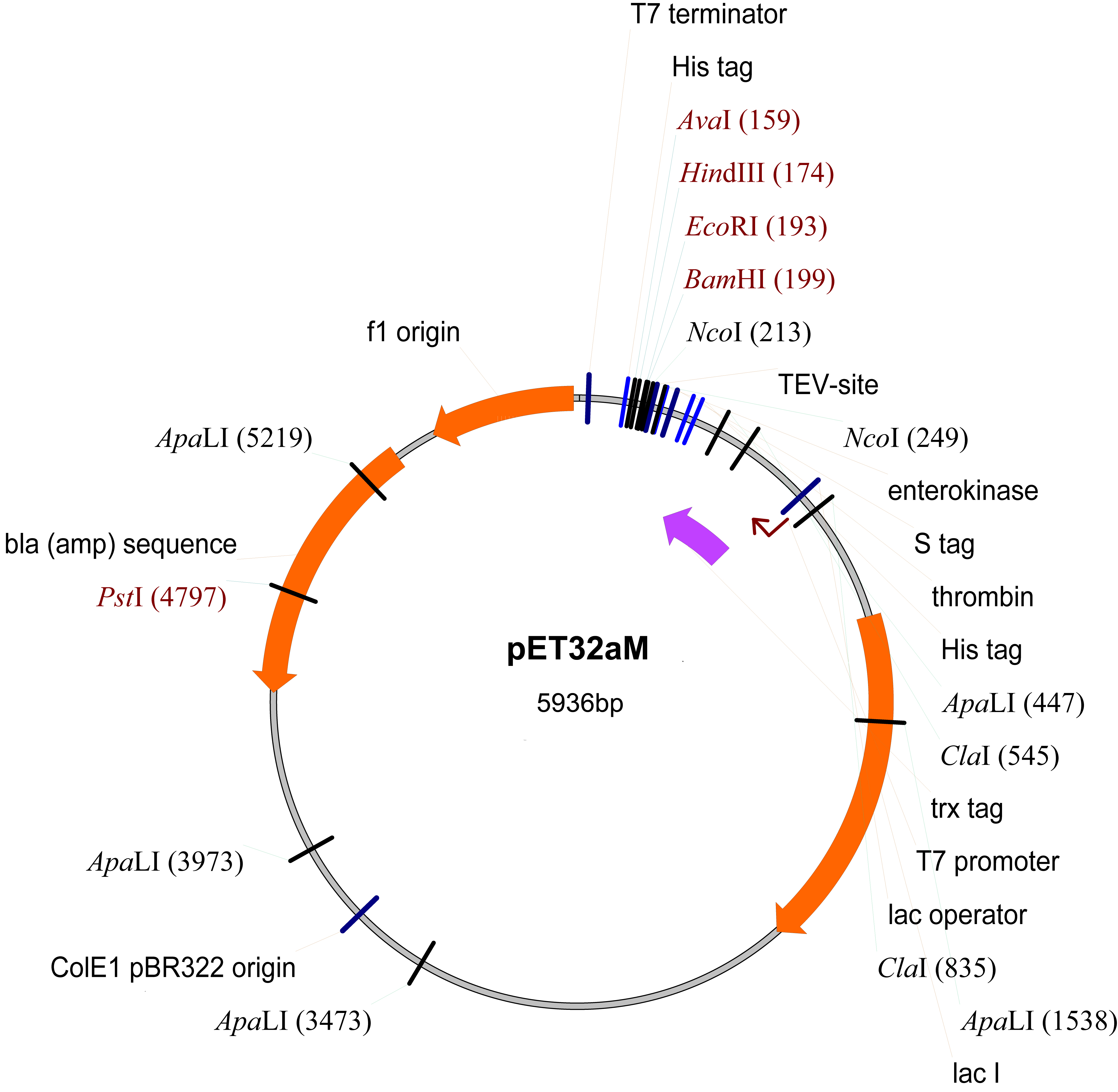
Figure 1. The map of pET32aM plasmid
Equipment
- 2 L flask
- Spectrophotometer device (Kebo instrument, model: UV-1100 )
- Sonicator (Scientz, model: IID )
- Refrigerated centrifuge (Eppendorf, model: 5810 R )
- Chromatography system (GE Healthcare, model: INV-907 )
Note: This product has been discontinued. - HiLoad 16/600 Superdex 200(GE Healthcare, catalog number: 28-9893-35 )
- Electrophoresis system (Liuyi, model: DYY-6D )
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Li, S., Zhao, Y. and Wu, Y. (2017). Expression and Purification of the GRAS Domain of Os-SCL7 from Rice for Structural Studies. Bio-protocol 7(3): e2122. DOI: 10.21769/BioProtoc.2122.
分类
植物科学 > 植物生物化学 > 蛋白质 > 分离和纯化
生物化学 > 蛋白质 > 表达
生物化学 > 蛋白质 > 结构
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link