- EN - English
- CN - 中文
Biochemical Analysis of Caspase-8-dependent Proteolysis of IRF3 in Virus-infected Cells
病毒感染细胞中IRF3被Caspase-8依赖性蛋白水解的生化分析
发布: 2016年11月20日第6卷第22期 DOI: 10.21769/BioProtoc.2018 浏览次数: 9271
评审: Yannick DebingAngela CoronaRosario Gomez-Garcia

相关实验方案
诱导型HIV-1库削减检测(HIVRRA):用于评估外周血单个核细胞中HIV-1潜伏库清除策略毒性与效力的快速敏感方法
Jade Jansen [...] Neeltje A. Kootstra
2025年07月20日 1553 阅读
Abstract
Interferon regulatory factor 3 (IRF3) is a transcription factor, which is critical for the antiviral response against a wide range of viruses (Hiscott, 2007; Ikushima et al., 2013). It gets activated in virus-infected cells via Toll like receptors (TLRs), RIG-I (retinoic acid inducible gene 1) like receptors (RLRs), cyclic GMP-AMP synthase (cGAS) – stimulator of interferon genes (STING), which are sensors of viral components in the cells (Chattopadhyay and Sen, 2014a; 2014b; Hiscott, 2007). IRF3 is a cytoplasmic protein, upon activation by virally activated sensors it gets phosphorylated, translocated to the nucleus and binds to the interferon-sensitive response element (ISRE) of the gene promoters to induce their transcription (Hiscott, 2007). IRF3 has other functions, including direct stimulation of apoptosis in virus-infected cells. In this pathway, the transcriptional activity of IRF3 is not required (Chattopadhyay et al., 2013b; Chattopadhyay et al., 2016; Chattopadhyay et al., 2010; Chattopadhyay and Sen, 2010; Chattopadhyay et al., 2011). These pathways are negatively regulated by host factors as well as by viruses. Our studies indicate that IRF3 can be proteolytically processed by caspase-8-dependent cleavage (Sears et al., 2011). A specific site in IRF3 is targeted by caspase-8, activated in RNA or DNA virus-infected and dsRNA-stimulated cells (Sears et al., 2011). The direct involvement of caspase-8 was confirmed by in vitro cleavage assay using recombinant proteins and in vivo by virus activated caspase-8. The proteolytic cleavage of IRF3 can be inhibited by chemical inhibition or genetic ablation of caspase-8. The cleavage of IRF3 removes the activated pool of IRF3 and thus can be used as a pro-viral mechanism (Figure 1). Using a C-terminally epitope-tagged human IRF3, we analyzed the cleavage of IRF3 in virus-infected cells. Moreover, we used recombinant proteins in vitro to conclude that IRF3 is a substrate of caspase-8 (Sears et al., 2011). In the current protocol, we have outlined a simple and detailed procedure to biochemically analyze the proteolysis of IRF3 in virus-infected cells and the specific role of caspase-8 in this process. 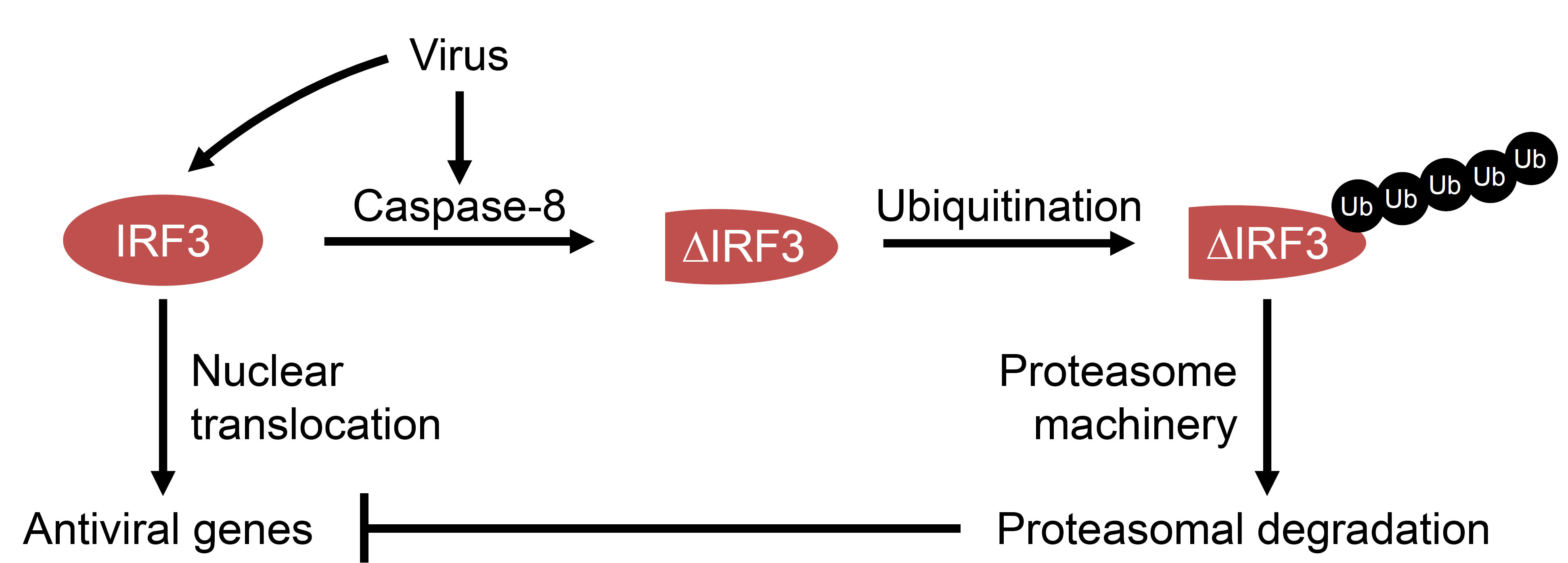
Figure 1. Model for virus-induced caspase-8-dependent proteolysis of IRF3. In virus-infected cells, IRF3 can be proteolytically cleaved by caspase-8, which gets activated during infection. The cleaved IRF3 is subjected to poly-ubiquitination (Ub) and degradation by proteasome machinery. The degradation of IRF3 inhibits dsRNA-induced antiviral gene induction.
Background
IRF3 functions as a transcription factor as well as a pro-apoptotic factor in virus-infected cells. The two properties of IRF3 determine its overall antiviral functions. Our studies have revealed a number of additional regulatory mechanisms of IRF3 for both of its functions. Using genetically modified cells and mice, we have shown how the two functional branches of IRF3 protect against virus infections (Chattopadhyay et al., 2013a; Chattopadhyay et al., 2016; Chattopadhyay et al., 2011). Dysregulated IRF3 activation is undesirable for normal physiological functions and, therefore, negative regulatory mechanisms are in place to control continuous activation of IRF3. In virus-infected cells, IRF3 can be proteolytically cleaved at a specific site by caspase-8, which gets activated by TLR or RLR signaling. The proteolytic cleavage of IRF3 prevents unregulated expression of dsRNA-dependent genes (Sears et al., 2011). We speculate that such mechanisms are used by viruses to promote their replication and to downregulate IRF3 responses. Therefore, the proteolysis of IRF3 represents a critical step, which both the viruses and the host can regulate to their favorable outcomes. This protocol provides a method to biochemically analyze the proteolysis of IRF3 in virus-infected cells.
Materials and Reagents
- Materials
- Autoradiography film (Denville Scientific, catalog number: E3012 )
- Tissue culture plates (Corning)
- 1.5 ml Eppendorf tubes (USA Scientific)
- Centrifuge tubes (Thermo Fisher Scientific)
- 26-gauge needle
- PVDF membranes
- Autoradiography film (Denville Scientific, catalog number: E3012 )
- Cells
- HT1080 fibrosarcoma cells
Note: These cells are maintained in DMEM containing 10% FBS, 100 international units of penicillin, 100 µg/ml streptomycin (complete DMEM). - 1080.10 cells (IRF3 overexpressing HT1080 cells [Elco et al., 2005])
Note: These cells are maintained in complete DMEM containing 1 µg/ml puromycin. - P2.1/IRF3 cells (P2.1 cells expressing C-terminally Flag-tagged human IRF3, Figure 2A)
- These cells were generated by lentiviral transduction of the P2.1 cells with cDNA of Flag-tagged human IRF3. After lentiviral transduction, the cells were selected in puromycin (1 µg/ml)-containing complete medium. Use the selected pool of cells for the experiments.
- HT1080 cells were described previously (Chattopadhyay et al., 2016) and the other cell lines (1080.10 and P2.1/IRF3) were generated in the authors’ laboratory.
- HT1080 fibrosarcoma cells
- Viruses
- Sendai virus (SeV) Cantell strain (Charles River laboratories) – this strain was originally obtained from ATCC (ATCC, catalog number: VR-907TM )
- Adenovirus Ad5 strain (a kind gift from Thomas Shenk, Princeton University, New Jersey)
- Sendai virus (SeV) Cantell strain (Charles River laboratories) – this strain was originally obtained from ATCC (ATCC, catalog number: VR-907TM )
- Reagents
- DMEM (Lerner Research Institute Central Cell Services, catalog number: 11-500p )
- PolyI:polyC (Poly [I:C]) (GE Healthcare, catalog number: 27-4732-01 )
- Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668030 )
- Opti-MEM I reduced serum medium (Thermo Fisher Scientific, InvitrogenTM, catalog number: 31985070 )
- Phosphate-buffered saline (PBS) (Lerner Research Institute Central Cell Services, catalog number: 123-1000p )
- Recombinant caspase-8 (EMD Millipore, catalog number: CC123 )
- Protein assay dye (Bio-Rad Laboratories, catalog number: 5000006 )
- 10x SDS-PAGE running buffer (AMRESCO, catalog number: 0783 )
- Antibodies:
anti-Flag (used at 1:2,500) (Sigma-Aldrich, catalog number: F3290 )
anti-IRF3 (dilution used at 1:10,000) (Active Motif, catalog number: 39033 )
anti-actin (used at 1:10,000 ) (Sigma-Aldrich, catalog number: A5441 )
HRP-conjugated secondary antibodies (used at 1:5,000) (Rockland Immunochemicals, catalog numbers: 611-103-121 , 611-103-122 ) - Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11550 )
- Proteasome inhibitor MG132 (EMD Millipore, catalog number: 474790 )
- Phosphatase inhibitor (PhosSTOP) (Roche Diagnostics, catalog number: 04906837001 )
- cOmplete EDTA-free protease inhibitor (Roche Diagnostics, catalog number: 11873580001 )
- Caspase inhibitors:
z-VAD (MBL, catalog number: 4800-520 )
z-WEHD (MBL, catalog number: BV-1100-3 )
z-IETD (MBL, catalog number: 4805-510 ) - Anti-Flag-agarose beads (Sigma-Aldrich, catalog number: A2220 )
- FLAG peptide (Sigma-Aldrich, catalog number: F3290 )
- SDS-PAGE loading buffer (Laemelli) (Bio-Rad Laboratories, catalog number: 161-0737 )
- Tris buffered saline with Tween-20 (TBS-T) (AMRESCO, catalog number: K873 )
- Nonfat dry milk (Bio-Rad Laboratories, catalog number: 1706404XTU )
- PierceTM ECL2 (Thermo Fisher Scientific, Fisher Scientific, catalog number: PI80196 )
- Tris buffer (pH 7.4) (Affymetrix, catalog number: 22639 )
- Sodium chloride (NaCl) (Affymetrix, catalog number: 75888 )
- Triton X-100 (Sigma-Aldrich, catalog number: T9284 )
- Sodium orthovanadate (NaVO4) (Sigma-Aldrich, catalog number: S6508 )
- Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: S7920 )
- β-glycerophosphate (Sigma-Aldrich, catalog number: G9422 )
- Sodium pyrophosphate (Na2H2P2O7) (Sigma-Aldrich, catalog number: P8135 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- CHAPS hydrate (Sigma-Aldrich, catalog number: C3023 )
- EDTA (Affymetrix, catalog number: 15694 )
- Glycerol (Affymetrix, catalog number: 16374 )
- 1,4-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 10197777001 )
- 10x transfer buffer (AMRESCO, catalog number: 0307 )
- Cell lysis buffer (see Recipes)
- Caspase-8 in vitro cleavage assay buffer (see Recipes)
- DMEM (Lerner Research Institute Central Cell Services, catalog number: 11-500p )
Equipment
- Tissue culture incubator (at 37 °C with 5% CO2) (Thermo Fisher Scientific)
- Biosafety cabinet
- Table top centrifuge (Eppendorf)
- Water bath (Benchmark)
- Heating block (at 95 °C) (Benchmark)
- SDS-PAGE and transfer apparatus (Bio-Rad Laboratories, model: Mini PROTEAN-II Cell )
Note: This equipment is no longer available at manufacturer. - Rocker (Benchmark)
- Rotator (Labnet)
- Refrigerator
- Autoradiography film processor (Kodak)
- Spectrophotometer
- Vortex (Thermo Fisher Scientific)
Software
- ImageJ (freely available from National Institutes of Health, https://imagej.nih.gov/ij/)
Procedure
文章信息
版权信息
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Subramanian, G., Pan, K., Chakravarti, R. and Chattopadhyay, S. (2016). Biochemical Analysis of Caspase-8-dependent Proteolysis of IRF3 in Virus-infected Cells. Bio-protocol 6(22): e2018. DOI: 10.21769/BioProtoc.2018.
- Sears, N., Sen, G. C., Stark, G. R. and Chattopadhyay, S. (2011). Caspase-8-mediated cleavage inhibits IRF-3 protein by facilitating its proteasome-mediated degradation. J Biol Chem 286(38): 33037-33044.
分类
微生物学 > 微生物-宿主相互作用 > 病毒
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link