- EN - English
- CN - 中文
Plant Sequence Capture Optimised for Illumina Sequencing
优化用于Illumina测序的植物序列捕获技术
发布: 2014年07月05日第4卷第13期 DOI: 10.21769/BioProtoc.1166 浏览次数: 16284
评审: Tie Liu
Abstract
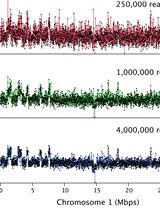
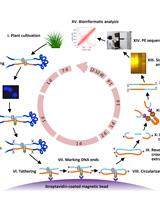
Plant Sequence Capture is used for targeted resequencing of whole exomes (all exons of a genome) of complex genomes e.g. barley and its relatives (Mascher et al., 2013). Sequencing and computing costs are significantly reduced since only the greatly enriched and gene-coding part of the barley genome is targeted, that corresponds to only 1-2% of the entire genome. Thus, applications such as genetic diversity studies and the isolation of single genes (“cloning-by-sequencing”) are greatly facilitated. Here, a protocol is provided describing the construction of shotgun DNA libraries from genomic barley DNA for sequencing on the Illumina HiSeq/MiSeq systems. The shotgun DNA sequencing libraries are hybridized to an oligonucleotide pool (Exome Library) encompassing the whole exome of barley. The Exome Library is provided as a liquid array containing biotinylated probes (Roche/NimbleGen). Subsequently, genomic shotgun DNA fragments hybridized to the Exome Library are affinity-purified using streptavidin coated magnetic beads. The captured library is PCR-amplified and sequenced using high-throughput short read sequencing-by-synthesis.
Materials and Reagents
- SYBR Gold (Life Technologies, catalog number: S11494 )
- UltraPure Agarose (Life Technologies, InvitrogenTM, catalog number: 16500-500 )
- PCR-grade water
- Tris (hydroxymethyl) aminomethane (Tris base) (multiple vendors)
- Tween 20 (Bio-Rad Laboratories, catalog number: 170-6531 )
- Ethylenediaminetetraacetic acid (EDTA) (multiple vendors)
- Ethanol (absolute) (analytical grade) (multiple vendors)
- 70 % (v/v) ethanol (analytical grade)
- Isopropanol (2-Propanol) (>99.5%) (multiple vendors)
- Acetic acid (glacial) (multiple vendors)
- 2x Phusion High-Fidelity PCR Master Mix (New England BioLabs, catalog number: F-531L )
- GeneRuler 50 bp DNA ladder (Thermo Fisher Scientific, catalog number: SM 0371 )
- QIAquick PCR Purification Kit (QIAGEN, catalog number: 28106 )
- Minelute PCR Purification Kit (QIAGEN, catalog number: 28006 )
- Agilent DNA 7500 Kit (Agilent Technologies, catalog number: 5067-1506 )
- Agilent High Sensitivity DNA Kit (Agilent, catalog number: 5067-4626 )
- SeqCap EZ Hybridization Kit [containing NimbleGen SC Wash Buffers (tubes 1, 2 and 3), Stringent Wash Buffer (tube 4), 2x SC Hybridisation Buffer (tube 5), Hybridisation Component A (tube 6) and the Bead Wash Buffer (tube 7)] (Roche Diagnostics, catalog number: 05634261001 )
- Sequence Capture Developer Reagent (Roche Diagnostics, catalog number: 06684335001 )
Note: This reagent was previously known as Plant Capture Enhancer (PCE) from Roche NimbleGen. - 6x loading dye (Thermo Fisher Scientific, Fermentas, catalog number: R 0611 )
- DNA away (Thermo Fisher Scientific, catalog number: 21-236-28 )
- Oligonucleotides (Sigma-Aldrich)
Note: All oligonucleotides were reverse phase cartridge purified and dissolved in PCR-grade water. To avoid cross-contaminations each oligonucleotide was purified using a fresh column. Sequences are listed in Table 1 in Supplementary Material. - Reagents for the generation of capture libraries:
- Illumina TruSeq DNA kit (Illumina, catalog number: FC-121-2001 , Box A) or
- TruSeq DNA PCR-free sample preparation kit (Illumina, catalog number: FC-121-3001 , Set A)
- If the DNA Illumina multiplex (IM) protocol is used, the following additional reagents (items 27- 41) are required.
- Illumina TruSeq DNA kit (Illumina, catalog number: FC-121-2001 , Box A) or
- T4 polynucleotide kinase (10 U/µl) (Thermo Scientific Fermentas, catalog number: EK0032 )
- T4 DNA polymerase (5 U/µl) (Thermo Scientific Fermentas, catalog number: EP006B )
- 10x Buffer Tango (Thermo Scientific Fermentas, catalog number: BY5 )
- dNTPs (25 nM each) (Thermo Scientific Fermentas, catalog number: R1121 )
- ATP (100 mM) (Thermo Fisher Scientific, catalog number: R0441 )
- T4 DNA ligase (5 U/µl) provided with 10x T4 DNA ligase buffer and 50% PEG 4000 (Thermo Scientific Fermentas, catalog number: EL0011 )
- 10x ThermoPol reaction buffer (New England Biolabs, catalog number: B9004S )
- Bst polymerase (8 U/µl) (large fragment) (New England Biolabs, catalog number: M0275L )
- 5x Phusion HF buffer (New England Biolabs, catalog number: B0518S )
- Phusion Hot Start Flex DNA polymerase (2 U/µl) (New England Biolabs, catalog number: M0535L )
- carboxyl-modified Sera-Mag Magnetic Speed-beads (Thermo Fisher Scientific, catalog number: 1182-9912 )
- PEG-8000 (Sigma-Aldrich, catalog number: 89510 )
- NaCl (Sigma-Aldrich, catalog number: S3014 )
- Agilent High Sensitivity DNA Kit (Agilent, catalog number: 5067-4626)
- Agilent DNA 7500 Kit (Agilent, catalog number: 5067-1506)
- TE (pH 8.0) (see Recipes)
- Resuspension Buffer (RSB) (see Recipes)
- 50x TAE (see Recipes)
- EBT (see Recipes)
- Test fragment for DNA Illumina multiplex (IM) libraries (see Recipes)
- Adapter mix P57 for IM libraries (see Recipes)
- MagNA beads for DNA clean-up (see Recipes)
Equipment
- Qubit 2.0 fluorometer (Starter Kit) [including instrument, assay tubes, dsDNA HS Assay and dsDNA BR Assay] (Thermo Fisher Scientific, catalog number: Q32871 ) or other picogreen-based dsDNA quantification devices
- AMPure XP Beads (Beckman Coulter, catalog number: A63882 )
- Streptavidin Dynabeads (M-270) (Life Technologies, InvitrogenTM, catalog number: 65306 )
- 1.5 ml tubes (multiple vendors)
- 0.2 ml PCR-tubes (multiple vendors)
- 96-well plates (Greiner Bio-One GmbH, catalog number: 652250 )
- Plastic seals for 96 well plate (Thermo Fisher Scientific, catalog number: AB-0558 )
- Covaris S220 AFA Ultrasonicator (LGC, catalog number: KBS-500217 ) and associated equipment such as microTUBE holder, chiller, software, computer etc.
- Snap-Cap microTUBES with AFA-fiber and pre-split septum (Covaris, catalog number: 520045 )
- Agilent 2100 Electrophoresis Bioanalyzer (Agilent, catalog number: G2939AA ) and associated material (e.g. computer)
- NanoDrop 2000 Spectrophotometer (VWR International, PeQlab, catalog number: 91-ND-2000 )
- HiSeq or MiSeq Illumina systems and associated materials (such as Sequencing-by-Synthesis reagents for 2x 100 cycles)
- Heating block (multiple vendors)
- Water Bath with external calibrated thermometer (multiple vendors)
- Microcentrifuge (16,000 x g) (multiple vendors)
- SpeedVac (multiple vendors)
- Thermocycler (multiple vendors)
Note: Throughout the protocol a “Bio Rad DNA Engine Tetrad 2 Peltier Thermal Cycler” was used. - Vortex Mixer (multiple vendors)
- DynaMag-2 Magnet (Life Technologies, InvitrogenTM, catalog number: 123-21D )
- DynaMag-96 Side Skirted Magnetic Particle Concentrator (MPC96) (Life Technologies, InvitrogenTM, catalog number: 120.27 )
- Agarose gel electrophoresis equipment and accessories [microwave, tray (15 x 15 cm), combs, power supply, UV-transilluminator, etc.] (multiple vendors)
- Disposable scalpels (multiple vendors)
- Dark Reader blue light transilluminator (Clare Chemical Research, catalog number: DR46B )
- Pipettes (2, 100, 200 and 1,000 µl, multiple vendors)
- Filter tips (multiple vendors)
Procedure
文章信息
版权信息
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Himmelbach, A., Knauft, M. and Stein, N. (2014). Plant Sequence Capture Optimised for Illumina Sequencing. Bio-protocol 4(13): e1166. DOI: 10.21769/BioProtoc.1166.
分类
植物科学 > 植物分子生物学 > DNA > DNA 测序
系统生物学 > 基因组学 > 外显子组捕获
分子生物学 > DNA > DNA 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link