往期刊物2024
卷册: 14, 期号: 17
细胞生物学
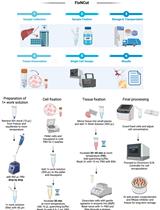
FixNCut: A Practical Guide to Sample Preservation by Reversible Fixation for Single Cell Assays
FixNCut:单细胞检测样本可逆固定保存实用指南
免疫学
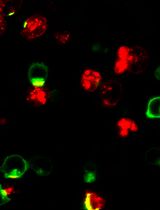
An Imaging-Based Assay to Measure the Location of PD-1 at the Immune Synapse for Testing the Binding Efficacy of Anti-PD-1 and Anti-PD-L1 Antibodies
用于测试抗PD-1和抗PD-L1抗体结合效力的基于成像的免疫突触PD-1定位检测方法
医学
Endothelin-1-Induced Persistent Ischemia in a Chicken Embryo Model
内皮素-1诱导的鸡胚持续性缺血模型
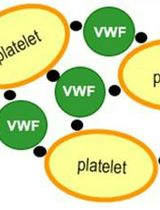
An Automated pre-Dilution Setup for Von Willebrand Factor Activity Assays
血管性血友病因子活性检测的自动化预稀释系统
微生物学
Multiplexed Microfluidic Platform for Parallel Bacterial Chemotaxis Assays
平行细菌趋化性检测的多重微流控平台
分子生物学
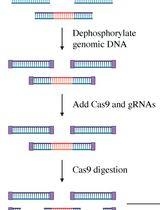
Single-Molecule Sequencing of the C9orf72 Repeat Expansion in Patient iPSCs
患者iPS细胞中C9orf72重复扩展的单分子测序
植物科学
Evaluating Mechanisms of Soil Microbiome Suppression of Striga Infection in Sorghum
评估土壤微生物组抑制高粱独脚金感染的机制
Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
拟南芥子房中轴与侧区的激光辅助显微切割及高通量RNA测序