往期刊物2022
卷册: 12, 期号: 22
生物工程
A Fast and Efficient Decellularization Method for Tissue Slices
一种快速、高效的组织切片去细胞方法
发育生物学
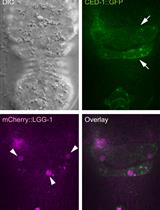
Monitoring the Recruitment and Fusion of Autophagosomes to Phagosomes During the Clearance of Apoptotic Cells in the Nematode Caenorhabditis elegans
监测秀丽隐杆线虫凋亡细胞清除过程中自噬体与吞噬体的募集与融合
药物发现
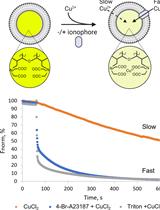
In vitro Assay to Evaluate Cation Transport of Ionophores
评估离子载体阳离子转运的体外检测方法
微生物学
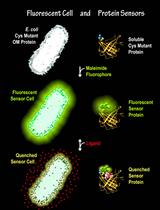
Fluorescent Binding Protein Sensors for Detection and Quantification of Biochemicals, Metabolites, and Natural Products
用于检测和量化生化物质、代谢物和天然产物的荧光结合蛋白传感器
Babesia duncani in Culture and in Mouse (ICIM) Model for the Advancement of Babesia Biology, Pathogenesis and Therapy
促进巴贝虫生物学、发病机制和治疗的巴贝虫培养和小鼠(ICIM)模型
Hypochlorite Stress Assay for Phenotypic Analysis of the Halophilic Archaeon Haloferax volcanii Using an Improved Incubation Method and Growth Monitoring
利用改进的培养方法和生长监测对嗜盐火山生菌进行表型分析的次氯酸盐胁迫试验
神经科学
Fluorescence Screens for Identifying Central Nervous System–Acting Drug–Biosensor Pairs for Subcellular and Supracellular Pharmacokinetics
用于识别亚细胞和超细胞药代动力学的中枢神经系统作用的药物-生物传感器配对的荧光筛选
In vitro Preparation of Homogenous Actin Filaments for Dynamic and Electrophoretic Light Scattering Measurements
用于动态和电泳光散射测量的均质肌动蛋白丝的体外制备
干细胞
Using BODIPY FL-Sphingolipid Analogs to Study Sphingolipid Metabolism in Mouse Embryonic Stem Cells
使用BODIPY FL-鞘脂类化合物研究小鼠胚胎干细胞中的鞘脂类代谢
系统生物学
Proteome Integral Solubility Alteration (PISA) Assay in Mammalian Cells for Deep, High-Confidence, and High-Throughput Target Deconvolution
在哺乳动物细胞中进行蛋白质组积分溶解度改变(PISA)测定,用于深度、高置信度和高通量目标解构
Efficient Generation of Genome-wide Libraries for Protein–ligand Screens Using Gibson Assembly
利用Gibson组装技术高效生成蛋白质配体筛选全基因组文库