Advanced Search
Cellular Retention Assay to Determine the Interaction Frequency of CD19-directed Chimeric Antigen Receptor (CAR) Engineered Cells against CD19+ Leukemic Cells
Published: Sep 5, 2019 DOI: 10.21769/BioProtoc.3358 Views: 3916
Edited by: Alka Mehra Reviewed by: Vemika Chandra
Abstract
Cancer recognition by chimeric antigen receptor T (CAR-T) cell is prerequisite for cancer killing to occur. Anomalies in the binding specificity of scFv in the CAR construct may prevent successful eradication of these cancer cells. In some cases, these anomalies (i.e., altered specificity) may cause deleterious effects such as on-target off-tumor toxicity or on-set CAR-T cell activation that may lead to life-threatening complications. We describe in this assay an easy, flexible and cheap way of analyzing target specificity of CAR-engineered cells toward cancer in the context of cell-to-cell interaction that can be used to screen other antigen-specific CARs. We are coining this test as cellular retention assay.
Keywords: Cell-to-cell interaction assayBackground
The encouraging clinical outcome endowed by CD19-directed chimeric antigen receptor T (CAR-T) cells in treating CD19+ hematologic malignancies motivated the development of more CAR-T platforms for therapeutic application in wide variety of cancer types including solid tumors and other types of liquid cancers (Newick et al., 2016; Park et al., 2016). Chimeric antigen receptors (CARs) are composed of 1) an antigen-specific scFv (single chain variable fragment) moiety found at the extracellular matrix and 2) T cell activating intracellular domain. CARs are constructed by fusing a cancer antigen-specific scFv of an antibody molecule with T cell-associated activation domain such as CD3 zeta only or in combination with certain co-stimulatory molecules such as CD137 (4-1BB), CD28 and/or in the presence of inducible cytokines (Figure 4A). While the intracellular region facilitates T cell-mediated cancer killing, the scFv domain of CAR performs the crucial role in the “search and attack” operation mounted by engineered T cells against cancer. Majority of pre-clinal workflow in CAR-T development relies heavily on characterizing the binding of scFv in unfused, CAR-independent context (such as in phage/ yeast display or produced from hybridoma technology) using several immuno-assays. Once the scFv is constructed and fused with the rest of the CAR expression cassette followed by T cell transduction or transfection, cytotoxicity and cancer killing in an in vitro co-culture challenge or xenogeneic mouse models are immediately performed without verifying the retained binding specificity of scFv in the CAR-fused form (Wang and Rivière, 2016; Levine et al., 2017; Vormittag et al., 2018).
Cancer cell recognition by CAR-T cell is a fundamental prerequisite for cancer killing to occur. Anomalies and defects in the binding specificity of scFv in the CAR-fused form may prevent the efficient anti-cancer activity of these engineered cells and may instead cause detrimental side effects which include on-target off-tumor cytotoxicity and on-set CAR-T activation. Due to the complexity of CAR cloning and construction, along with the intricate and complicated antigen recognition mechanisms contributed by cancer pro-survival strategies, the poor and expensive structural elucidation within cellular environment and the unpredicted high toxicity rate of CAR-T therapy contributed by undesirable scFv characteristics (De los Santos and Bernal, 2018), the analysis of retained antigen specificity of scFv in the CAR-fused form is very crucial in the pre-clinal workflow of CAR-T development.
In this assay, we demonstrate the retainment of target specificity of CAR-engineered cell with cancer antigen in a cell-to-cell interaction assay (cellular retention assay). HEK293 cells are engineered to express the CD19-specific CAR (serving as CAR-engineered cells). An in-house established leukemic cell line established from cancer blood biopsy (Cayrefourcq et al., 2015), AMLK cells, was used as a cancer target model, bearing the CD19 antigen at the current passage. This cellular retention assay measures the ability of cell-to-cell mediated substrate-anchorage retention as a measure of CAR scFv binding to target cancer antigen as illustrated in Figure 1 below.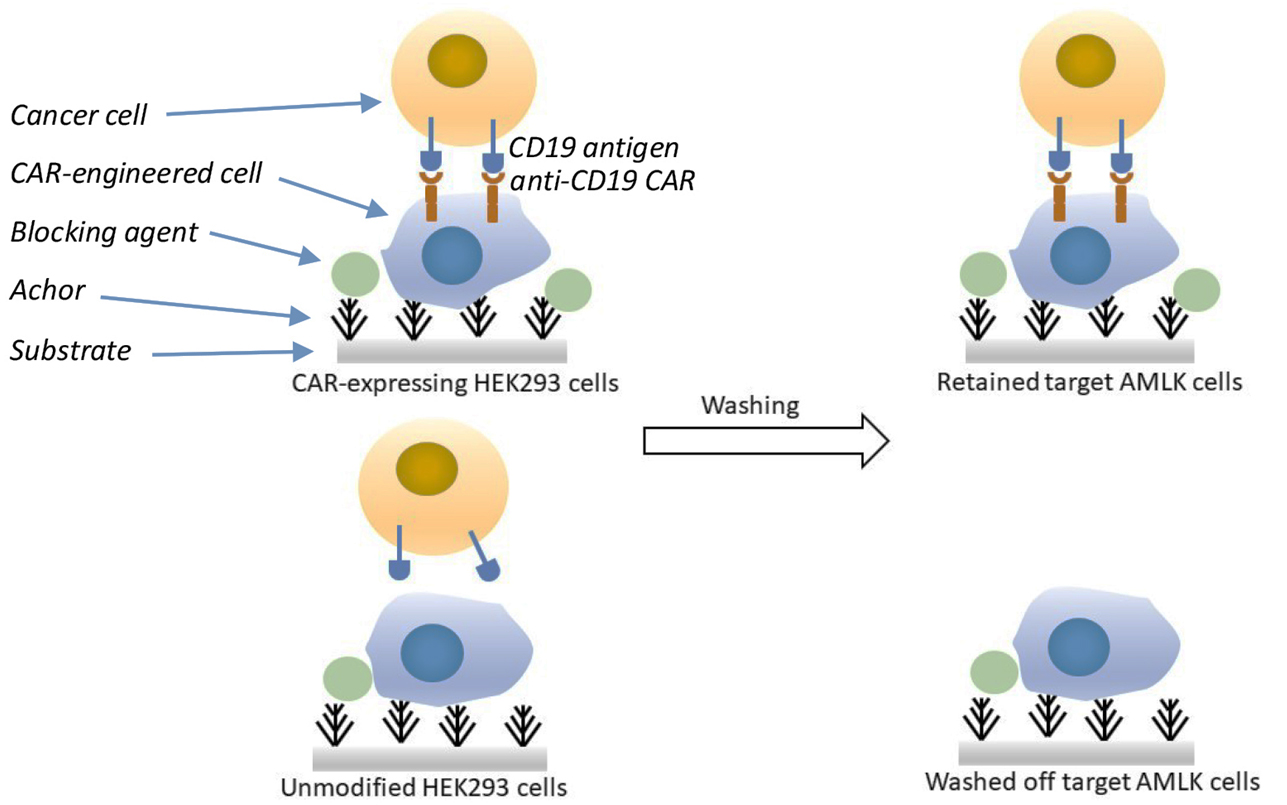
Figure 1. Cellular retention assay model to determine cell-to-cell mediated recognition of CD19 CAR engineered cells with CD19+ leukemic AMLK cells
Materials and Reagents
- Cell culture expansion
- 75 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353110)
- 25 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353108)
- 15 ml conical tube (Fisher Scientific, FisherbrandTM, catalog number: 05-539-5)
- Serological pipette 5 ml (Corning, StripetteTM, catalog number: 4487)
- Serological pipette 10 ml (Corning, StripetteTM, catalog number: 4488)
- HEK293 cells (ATCC, catalog number: CRL-1573)
- Target cancer cells (AMLK acute myeloid leukemia cells, Globetek Science Foundation Inc, Makati City, Philippines)
- Dulbecco's Modified Eagle Medium (Mediatech, catalog number: 10-013-CV)
- RPMI 1640 (Thermo Fisher Scientific, GibcoTM, catalog number: 11875093)
- Heat inactivated fetal bovine serum (Sigma-Aldrich, catalog number: F4135)
- Phosphate buffered saline, 1x (Mediatech, catalog number: 21-031-CV)
- Sodium bicarbonate, 7.5% (Thermo Fisher Scientific, Gibco, catalog number: 25080094)
- Media 1 (see Recipes)
- Media 2 (see Recipes)
- Lipofection and CD19-CAR expression in HEK293 cells
- 6-well plates, flat-bottom (Corning, Falcon®, catalog number: 353046)
- 15 ml conical tube (Fisher Scientific, FisherbrandTM, catalog number: 05-539-5)
- 1.5 ml Eppendorf microcentrifuge tubes (Sigma-Aldrich, catalog number: T9661-500EA)
- Micropipette tips [Thermo Fisher Scientific, catalog numbers: TF102-20-Q (0.1-10 µl); TF140-200-Q (20-200 µl); TF112-1000-Q (100-1,000 µl)]
- Phosphate buffered saline, 1x (Mediatech, catalog number: 21-031-CV)
- CAR construct (anti-CD19 scFv-CD28-CD137-CD3z) and empty mammalian expression vector
- Lipofectamine 2000® reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668-027)
- Opti-MEMTM I reduced serum medium (Thermo Fisher Scientific, GibcoTM, catalog number: 11058021)
- Membrane protein extraction kit such as ReadyPrepTM Extraction Kit- Membrane I (Bio-Rad, catalog number: 163-2088)
- Qubit protein assay quantification kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q33211)
- Laemmli sample buffer, 2x (Bio-Rad, catalog number: 1610737)
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250-100ML)
- NuPAGETM PAGE gels (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0336BOX)
- Precision Plus ProteinTM All Blue Prestained Protein Standards (Bio-Rad, catalog number: 1610373)
- Molecular biology grade water (Fisher Scientific, HyCloneTM, catalog number: SH3053803)
- MOPS (Fisher Scientific, Fisher BioReagentsTM, catalog number: BP308-100)
- Tris (Fisher Scientific, Fisher BioReagentsTM, catalog number: BP152-500)
- EDTA (Fisher Scientific, Fisher ChemicalTM, catalog number: S316-212)
- SDS (Fisher Scientific, Fisher BioReagentsTM, catalog number: BP166-100)
- Coomasie Brilliant Blue R250 (Thermo Fisher Scientific, catalog number: 20278)
- Glacial acetic acid (RCI Labscan, catalog number: AR1002-P2.5L)
- Methanol (RCI Labscan, catalog number: AR1115-P4L)
- MOPS-SDS running buffer (see Recipes)
- Coomasie protein stain (see Recipes)
- Destaining solution (see Recipes)
- Cellular retention assay
- 24-well plates, flat-bottom (Corning, Falcon®, catalog number: 353047)
- 1.5 ml Eppendorf microcentrifuge tubes (Sigma-Aldrich, catalog number: T9661-500EA)
- Micropipette tips [Thermo Fisher Scientific, catalog numbers: TF102-20-Q (0.1-10 µl); TF140-200-Q (20-200 µl); TF112-1000-Q (100-1,000 µl)]
- 15 ml conical tube (Fisher Scientific, FisherbrandTM, catalog number: 05-539-5)
- Countess cell counting chamber slides (Thermo Fisher Scientific, InvitrogenTM, catalog number: C10228)
- Poly-L-lysine (ScienceCell, catalog number: 0413)
- Bovine serum albumin (Fisher Scientific, Fisher BioreagentsTM, catalog number: BP9703100)
- Anti-CD19 antibody (Santa Cruz Biotechnology, IgG1 kappa light chain F-3 clone, catalog number: sc-373897)
- Non-specific antibody, isotype control (Santa Cruz Biotechnology, IgG1 kappa light chain AT80 clone, catalog number: sc-58982)
- Trypan blue stain, 0.4% (Thermo Fisher Scientific, InvitrogenTM, catalog number: T10282)
- Phosphate buffered saline, 1x (Mediatech, catalog number: 21-031-CV)
Equipment
- Electric pipet controller (Fisher Scientific, FisherbrandTM, catalog number: 14-955-202)
- Pipettes
- Cell culture CO2 incubator (Nuaire, model number: NU-5100E)
- Cell processing centrifuge (Hermle, Universal Centrifuge, model number: Z 366)
- Microliter centrifuge (Thermo Fisher Scientific, SORVALL Legend Micro 21, model number: D-37520)
- BioSafety cabinet
- Inverted microscope (ZEISS, Primo Vert, catalog number: 12-070-466)
- PAGE system (Thermo Fisher Scientific, XCell SureLockTM Mini Cell, catalog number: EI0001)
- Countess I automated cell counter (Thermo Fisher Scientific, InvitrogenTM, catalog number: C10227)
Software
- Office Excel (2003 or later versions, Microsoft) or any spreadsheet software
Procedure
Category
Cancer Biology > Tumor immunology > Cancer therapy > Cell transfer therapy
Cancer Biology > Oncogenesis > Leukemogenesis
Cell Biology > Cell-based analysis > Cell-to-cell interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
