Advanced Search
Ty1 Retrotransposition Frequency Assay Using a Chromosomal Ty1his3AI or Ty1kanMXAI Element
Published: Sep 5, 2018 DOI: 10.21769/BioProtoc.3004 Views: 4833
Abstract
Here I describe a simple genetic assay to determine the frequency of retrotransposition of a single chromosomal Ty1 element that is marked with the retrotransposition indicator gene, his3AI or kanMXAI. The assay is used to determine the effect of mutations or environmental conditions on the frequency of Ty1 retrotransposition in the yeast, Saccharomyces cerevisiae.
Keywords: Ty1Background
Ty1 is a long terminal repeat (LTR) retrotransposon that is structurally and evolutionarily related to retroviruses. Retrotransposition of Ty1 occurs when Ty1 RNA is reverse transcribed within cytoplasmic virus-like particles, resulting in synthesis of a cDNA that is transported back to the nucleus and integrated into the genome of its host (Boeke et al., 1985; Garfinkel et al., 1985). The integrated Ty1 element consists two LTRs in the same orientation that flank two open reading frames: GAG, which encodes a structural protein that forms the virus-like particle and binds Ty1 RNA, and POL, which encodes three enzymatic proteins- protease, reverse transcriptase and integrase (Figure 1). Ty1 elements are the most active and abundantly transcribed of all five families of LTR-retrotransposons in S. cerevisiae. Ty1 has been shown to be regulated by hundreds of genes, several different environmental conditions and various DNA damaging agents (Curcio et al., 2015). To measure the relative frequencies of Ty1 retrotransposition in different genetic backgrounds and under different environmental conditions, a chromosomal Ty1 element under the control of the native promoter was marked with a retrotransposition indicator gene (Figure 1). A retrotransposition indicator gene is a selectable marker gene whose coding sequence is interrupted by an intron in the opposite orientation to transcription (Figure 1). When placed within the Ty1 retrotransposon such that Ty1 and the marker gene are in opposite transcriptional orientations, the intron is spliced out of the Ty1 transcript but cannot be spliced out of the marker gene transcript. When the spliced Ty1 transcript undergoes retrotransposition, however, it recreates a functional marker gene in the transposed copy of the element. The fraction of viable cells in which the selectable marker gene is detected phenotypically is the retrotransposition frequency. This assay is a modification of that first described by Curcio and Garfinkel in 1991.
The specific assay conditions provided here are for derivatives of strain BY4741 containing a chromosomal Ty1 element that is marked with the retrotransposition indicator gene, his3AI or kanMXAI (Curcio and Garfinkel, 1991, Bryk et al., 2002). Cells that sustain retrotransposition of Ty1his3AI harbor a Ty1HIS3 element and are His+ prototrophs (i.e., capable of growth on medium lacking histidine; Figure 1). Cells that sustain retrotransposition of Ty1kanMXAI harbor a Ty1kanMX element and are resistant to G418.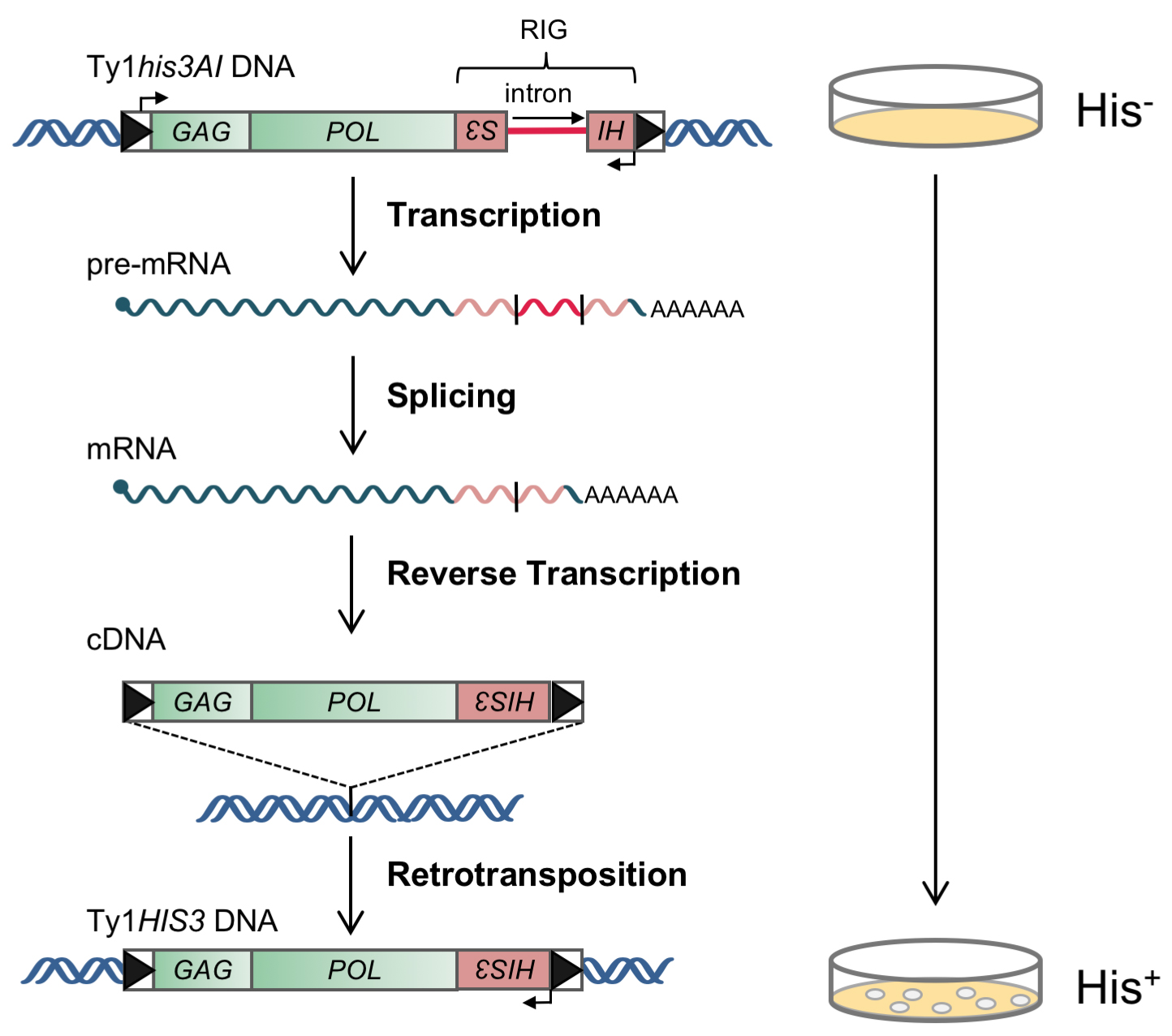
Figure 1. Genetic assay for detection of cells that sustain a retrotransposition event of the chromosomal Ty1his3AI element. The Ty1 element consists of long terminal repeats (boxed arrowheads) flanking a GAG and a POL open reading frame. The his3AI indicator gene, which consists of the HIS3 marker gene interrupted by an intron in the antisense orientation, is inserted downstream of POL. Ty1his3AI RNA is transcribed, and the intron within the HIS3 open reading frame is spliced out. The spliced RNA is then subject to reverse transcription during the process of retrotransposition. The resulting Ty1HIS3 cDNA lacks the intron and contains functional HIS3 gene sequences. Integration of the cDNA into the host genome renders the cell His+. (Adapted from Curcio et al., 2015)
Materials and Reagents
- Pipette tips for P10, P100 and P1000 micropipettes
- Sterile flat wood toothpicks for obtaining single colonies (VWR, catalog number: 470146-908)
Manufacturer: Regional Distributors, catalog number: 159864 . - Sterile 12 x 75 mm polypropylene tubes with caps (4 per strain tested) (Evergreen Scientific, CaplugsTM, catalog number: 222-2367-080 )
- Borosilicate glass 20 mm x 150 mm culture tubes (Sigma-Aldrich, catalog number: C1048 ) with closure caps (Sigma-Aldrich, catalog number: C1298 ), sterilized (1 per strain tested)
- Sterile 1.5 ml microfuge tubes (4 per strain tested)
- Petri dishes (Fisher Scientific, FisherbrandTM, catalog number: FB0875713 )
- Sterile Rattler Plating Glass Beads, 4.5 mm (Zymo Research, catalog number: S1001 )
- Strains
This assay is performed using derivatives of strain BY4741 that contain a Ty1his3AI[∆1] element, which has the “∆1” version of the his3AI gene. Ty1his3AI[∆1] must be used in strains such as BY4741 that have a his3∆1 allele rather than a complete deletion of the HIS3 gene. DNA recombination between his3AI[∆1] and his3∆1 cannot yield a functional HIS3 allele, so all His+ prototrophs that arise are due to the presence of a retrotransposed Ty1HIS3 element. Strains containing the chromosomal Ty1his3AI[∆1]-3114 element include JC3212 and JC3787 (Mou et al., 2006). The genotypes are:
JC3212 MATa his3∆1 leu2∆0 met15∆0 ura3∆0 Ty1his3AI[∆1]-3114
JC3787 MATα his3∆1 leu2∆0 met15∆0 ura3∆0 Ty1his3AI[∆1]-3114
A derivative of strain BY4741 containing a single chromosomal Ty1kanMXAI element was described recently (Salinero et al., 2018). This strain has the following genotype:
JC6464 MATa his3∆1 leu2∆0 met15∆0 ura3∆0 Ty1kanMXAI-6464 - YPD agar (Sunrise Science, catalog number: 1876-500 ), sterilized, in Petri dishes (4 plates per strain tested)
- SC Complete-His Powder (Sunrise Science, catalog number: 1481-100 )
- Agar, Yeast Culture Grade (Sunrise Science, catalog number: 1910-1KG )
- G418 Sulfate (GeneticinTM Selective Antibiotic) (Thermo Fisher Scientific, catalog number: 11811023 )
- Sterile YPD broth (Sunrise Science, catalog number: 1875-019 )
- Sterile deionized water (Rockland Immunochemicals, catalog number: MB-009-1000 )
- Ethyl alcohol (ethanol), 190 proof, non-denatured (Sigma-Aldrich, catalog number: E7148 )
- SC-HIS agar plates (see Recipe 1), or, Sterile YPD + 200 μg/ml G418 agar plates (see Recipe 3)
Note: A total of 8 plates (of either type) is needed per strain tested. - 200 mg/ml G418 stock solution (see Recipe 2)
Equipment
- P10, P100 and P1000 micropipettes
- Pipet-Aid
- Tissue Culture Rotator (Fisher Scientific, FisherbrandTM, catalog number: 14-251-250 )
- Rotator drum (Fisher Scientific, FisherbrandTM, catalog number: 14-251-251 )
- Benchtop centrifuge that accommodates 12 x 75 mm tubes in a swinging bucket rotor (Eppendorf, Centrifuge, model: 5810 with swing out rotor S-4-104, catalog number: 5820740000 )
- Microcentrifuge (Fisher Scientific, FisherbrandTM, model: accuSpinTM Micro 17, catalog number: 13-100-675 )
- Vortex Mixer (Thermo Fisher Scientific, MaxiMix®, model: M16715Q , catalog number: 12-815-50)
- Autoclave
Procedure
Category
Microbiology > Microbial genetics > Retrotransposition
Cell Biology > Cell-based analysis > Colony formation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

