Advanced Search
Growth of Chlamydomonas reinhardtii under Circadian Conditions
Published: Aug 20, 2018 DOI: 10.21769/BioProtoc.2982 Views: 5650
Edited by: Adam Idoine Reviewed by: Takuya Matsuo
Abstract
The green biflagellate unicellular alga Chlamydomonas reinhardtii serves as a model to study fundamental biological processes such as the structure and function of flagella or light-driven processes including photosynthesis, its behavioral responses, life cycle and circadian clock. Light-dark, as well as temperature cycles, are major Zeitgebers to entrain the algal circadian clock. In C. reinhardtii, several processes are under circadian control and many clock-controlled genes and/or proteins have been found in the past decades as well as components of the endogenous oscillator. Here, we describe a protocol for the growth of C. reinhardtii for the synchronization and analysis of its circadian clock.
Keywords: Chlamydomonas reinhardtiiBackground
In the past years, several clock components of C. reinhardtii have been identified and their function has been studied (for reviews, see Schulze et al., 2010; Matsuo and Ishiura, 2011; Noordally and Millar, 2015; Ryo et al., 2016; Kottke et al., 2017). In this article, we will present growth conditions for studying circadian control in C. reinhardtii. Therefore, we introduce the chronobiology nomenclature used for the growth of the algal cells under diurnal and circadian conditions (Figure 1).
At first, the circadian clock is synchronized by a light-dark cycle of 12-h light and 12-h dark, known as LD 12:12 at constant temperature. At LD0, light is turned on, and at LD12 it is switched off. LD6 thus defines the middle of the day and LD18 the middle of the night. A rhythm observed under LD conditions is called diurnal. Time measurement under diurnal conditions goes from LD0 to LD24. The next day is defined in the same way (LD0 to LD24). To find out if this rhythm is controlled by the circadian clock, the cells have to be released under so-called “free running conditions” with constant light and temperature where a circadian rhythm will continue with a period of about 24 h (Wagner and Mittag, 2009; Boesger et al., 2014). The cells are released at the end of the dark period (LD24) to constant conditions (Figure 1). Therefore, dim light (LL) is often used for C. reinhardtii, but if effects of specific light pulses are necessary as for the rhythm of photoaccumulation or for phase shifting the circadian clock, constant darkness (DD) is also used. For the rhythm of photoaccumulation (also described as rhythm of phototaxis), specialized set-ups and needs are necessary that differ depending on the home-made instrumental device (Mergenhagen, 1984; Gaskill et al., 2010; Forbes-Stovall et al., 2014; Müller et al., 2017). These are not further described in the current protocol.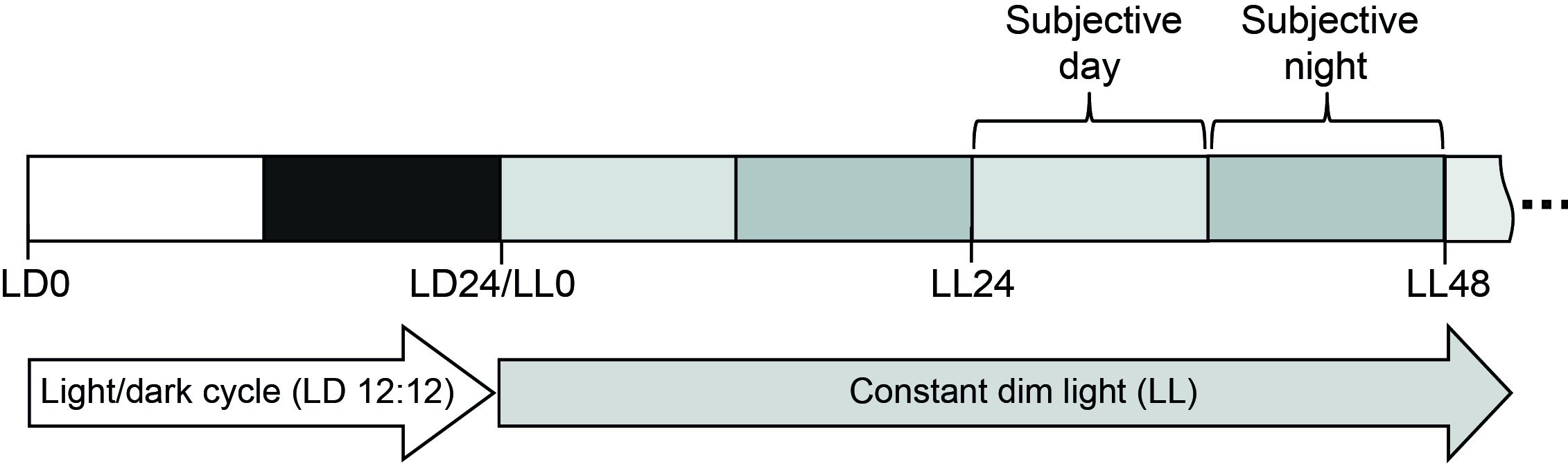
Figure 1. Light conditions for investigating circadian rhythms in C. reinhardtii
Under circadian conditions, time measurement starts at LL0 and continues with the number of hours under which the organism has been put under circadian conditions. For example, LL48 means that the organism was for two days under constant conditions. LL30 symbolizes the middle of subjective day and LL42 the middle of subjective night. Subjective day (or day phase) and subjective night (night phase) are commonly used terms for free running conditions in chronobiology. Since transients may occur upon transfer to constant conditions, circadian rhythms are usually measured after the organism has been exposed for at least 12 h to constant conditions, and often after exposure for 24 h.
Materials and Reagents
- Petri dishes
- Aluminum foil
- Autoclave tape
- Whatman® Prepleated Qualitative Filter Paper (GE Healthcare, catalog number: 1201-320 )
- Sterile tooth picks
- Indicator paper pH-Fix 0-14 (Machery-Nagel, catalog number: 92110 )
- Cotton plug
- Nunc®-flasks (NuncTM EasYFlaskTM 25cm2 with filter cap, gamma irradiated) (Thermo Fisher Scientific, catalog number: 156367 )
- Chlamydomonas reinhardtii cells
- Wild-type strain SAG 73.72 cells
- Double-distilled water (ddH2O; conductivity ≤ 0.1 μS/cm)
- NH4Cl
- CaCl2•2H2O
- MgSO4•7H2O
- K2HPO4
- KH2PO4
- Na2EDTA
- H3BO3
- FeSO4•7H2O
- 20% KOH
- ZnSO4•7H2O
- MnCl2•4H2O
- CoCl2•6H2O
- CuSO4•5H2O
- (NH4)6Mo7O24•4H2O
- Tris [Tris(hydroxymethyl)-aminomethane]
- Lugol's solution (iodine-potassium iodide solution; Merck, catalog number: 1092611000 )
- Liquid nitrogen
Caution: Extremely cold liquid (-196 °C); it may cause cryogenic burns or injury and displaces oxygen, which could lead to rapid suffocation in closed rooms; transport and store it always in containers designed for cryogenic liquids; handle it with special devices using protective clothing, cold insulating gloves and a face shield. - TAP salt solution (see Recipes)
- 1 M potassium phosphate buffer (see Recipes)
- Hutner's trace elements (see Recipes)
- Tris-acetate-phosphate (TAP) medium (see Recipes)
Equipment
- 1 L beaker
- Heater
- Erlenmeyer-flasks
- Magnetic stirrer
- Autoclave
- Sterile bench
- Culture room
- Laboratory sample shaker
- pH-electrode
- Centrifuge
- Neubauer improved counting chamber (Marienfeld, catalog number: 0640010 )
Procedure
Category
Plant Science > Phycology > Physiology
Microbiology > Microbial cell biology > Cell-based analysis
Plant Science > Plant physiology > Circadian clock
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
