Advanced Search
High-throughput YO-PRO-1 Uptake Assay for P2X7 Receptors Expressed in HEK Cells
Published: Jul 20, 2018 DOI: 10.21769/BioProtoc.2943 Views: 7770
Abstract
P2X7 receptors are extracellular ATP-gated ion channels that play broad physiological and pathological roles in animals (Sluyter, 2017). Activation of P2X7 receptors lead to the opening of membrane channels permeable for small cations like Na+ and Ca2+ as well as fluorescent dyes such as YO-PRO-1 (Alves et al., 2014). Taking advantage of this dye-permeability, YO-PRO-1 uptake assays have been widely used to probe P2X7 receptor activity (Surprenant et al., 1996; Rassendren et al., 1997; Karasawa et al., 2017). Here we describe a step by step protocol for a high-throughput YO-PRO-1 uptake assay using HEK293 cells expressing P2X7 receptors. This 3-day protocol is particularly suited for examining effects of small molecules and mutations on P2X7 receptor function. This protocol was adapted from our previously published paper (Karasawa and Kawate, 2016).
Keywords: P2X7Background
The P2X7 receptor opens a membrane channel permeable to cations (Surprenant et al., 1996; Sluyter, 2017). While electrophysiology remains the gold standard for quantifying P2X7 receptor activity, this specialized technique may not be readily available. In addition, electrophysiology is not suited for high-throughput screening, as it requires a substantial amount of time and manipulation for each recording. Uptake of a fluorescent molecule, therefore, has been a widely used alternative, especially for screening multiple conditions/mutants (Cankurtaran-Sayar et al., 2009; Qu et al., 2011). One of the most commonly used fluorescent molecules is a monomeric cyanine nucleic acid stain, YO-PRO-1. This fluorescent molecule increases its green fluorescence upon binding to nucleic acids in the cell. Because YO-PRO-1 directly permeates the P2X7 membrane pore, activity of this receptor can be studied by following the changes in green fluorescence (Karasawa et al., 2017). Typically, YO-PRO-1 uptake is studied using a fluorescent microscope, which allows time-dependent quantification of regions-of-interest (Surprenant et al., 1996; Rassendren et al., 1997). A major challenge, however, is to retain information about the activation kinetics while maintaining a higher throughput. To overcome this issue, we have developed a YO-PRO-1 uptake method using a fluorescent plate reader. By quickly measuring the total fluorescence from each well, our method enables data collection every 70-80 sec/well. While this is not near the sampling frequency of a low-throughput single cell measurement (e.g., electrophysiology or fluorescent microscopy), it is sufficient for quantifying the initial rate of YO-PRO-1 uptake. Our newly developed method described in this protocol, therefore, is an efficient approach for screening hundreds of conditions based on P2X7 activation kinetics.
Materials and Reagents
Note: Materials and Reagents are stored at room temperature unless otherwise noted.
- Generic pipette tips (VWR, catalog numbers: 613-0741 , 613-2133 , 613-0746 )
- Cryogenic Storage Vials 2 ml, Sterile (Fisher Scientific, catalog number: 10-500-26 )
- CELLSTAR serological pipettes 2, 5, 10 ml (Greiner Bio One International, catalog numbers: 710107 , 606107 , 607107 )
- Neptune Microcentrifuge Tubes with Attached Flat Caps 1.5 ml (Biotix, catalog number: 4445.X )
- Falcon® Tissue Culture Dishes (Corning, catalog number: 353003 )
- CELLSTAR 6-Well Multiwell Culture Plates (Greiner Bio One International, catalog number: 657160 )
- Corning® BioCoatTM 96-Well Multiwell Plates with Poly-D-Lysine (Corning, catalog number: 356640 )
- Multiwell Cell Culture 96-Well Plates (VWR, catalog number: 10062-900 )
- HEK-293 cells (ATCC, catalog number: CRL-1573 ) (stored in liquid nitrogen)
- pIM2 vector (developed in the Kawate lab; Figure 1) harboring a P2X7 receptor gene (stored in -20 °C freezer)
Note: In this protocol, panda P2X7 gene (NCBI Reference Sequence: XP_002913164.2) is used. - MAX EfficiencyTM DH5αTM competent cells (Thermo Fisher Scientific, InvitrogenTM, catalog number: 18258012 ) (Stored in -80 °C freezer)
- E.Z.N.A. Plasmid Mini Kit (Omega Bio-tek, catalog number: D6942-02 )
- Liquid nitrogen (Airgas, catalog number: NI 180LT230 )
- DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: 11995065 ) (stored in 4 °C fridge)
- FBS (Atlanta Biologicals, catalog number: S11150H ) (Stored in -80 °C freezer)
- Ultra Pure Grade Carbon Dioxide (Airgas, catalog number: CD UP200 )
- 100% Pure Ethanol (Decon Labs, catalog number: V1016TP )
- 2-Propanol (Avantor Performance Materials, catalog number: 9095-33 )
- DPBS/Modified (GE Healthcare, catalog number: SH30028.02 )
- HycloneTM Trypsin 0.25% (1x) (GE Healthcare, catalog number: SH30042.02 ) (stored in 4 °C fridge)
- YO-PROTM-1 Iodide (491/509) (Thermo Fisher Scientific, catalog number: Y3603 ) (stored in -20 °C freezer)
- jetPRIME (Polyplus-transfection, catalog number: 114-15 ) (stored in 4 °C fridge)
- Sodium Chloride (NaCl) (Fisher Scientific, catalog number: S641-212 )
- Gentamicin (Thermo Fisher Scientific, GibcoTM, catalog number: 15710064 )
- Hydrochloric acid (HCl) (VWR, catalog number: BDH7204-4 )
- Adenosine 5’-triphosphate disodium salt hydrate (Sigma-Aldrich, catalog number: A7699 ) (stored in a -20 °C freezer)
- HEPES (Fisher Scientific, Fisher BioreagentsTM, catalog number: BP310-1 )
- D-(+)-Glucose (Sigma-Aldrich, catalog number: G5767 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- Calcium Chloride Dihydrate (CaCl2·2H2O) (Fisher Scientific, catalog number: C79-500 )
- JNJ 47965567 (Tocris Bioscience, catalog number: 5299 ) (stored in 4 °C fridge)
- DMEM-FBS (see Recipes)
- Assay buffer (see Recipes)
- YO-PRO-1 Solution (see Recipes)
- 100 mM ATP stock (see Recipes)
- 100 mM Inhibitor stock (JNJ) (see Recipes)
Equipment
- 500 ml beaker
- Original Pipet-Aid pipette controller (Drummond Scientific, catalog number: 4-000-110 )
- GilsonTM PIPETMANTM M Multichannel Pipettes (Gilson, catalog number: F81027 )
- ErgoOne 30-300 μl 12-Channel (USA Scientific, catalog number: 7112-3300 )
- Mettler Toledo-XP204-Analytical Balance (Mettler-Toledo International, model: XP204S/M , catalog number: 11130054)
- Magnetic stir bars
- Baker SterilGARDR® II Biological safety Cabinet SG-600 (The Baker Company, model: SterilGARDR II SG600 )
- NanoDrop2000 (Thermo Fisher Scientific, ThermoScientific, model: NanoDropTM 2000 , catalog number: ND-2000)
- VWR® Ultra Low Temperature Upright Freezers and Freezer (VWR, model: 5656 )
- IsotempTM digital-control water baths (Fisher Scientific, model: Model 202 )
- Shel Lab 2350-T CO2 Water Jacketed Incubator (Sheldon Manufacturing, catalog number: 9150933 )
- MALGENECryo 1 °C Freezing Container (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 5100-0001 )
- Hemocytometer (Daigger Scientific, catalog number: EF16034F )
- Milli-Q® Reference Water Purification System (EMD Millipore, catalog number: Z00QSV0WW )
- SevenEasy pH meter (Mettler-Toledo International, catalog number: 51302819 )
- Gravity and vacamatic sterilizers (Amscope, model: EAGLE® SERIES 3000 , catalog number: 213415)
- Refrigerator (PHC, model: MPR-1411 )
- SANYO Biomedical freezer (SANYO, model: MDF-U333 )
- Steadystir digital S56 (Fisher Scientific, catalog number: 14-359-756 )
- Ice maker (Hoshizaki America, model: F-500BAF )
- Ice Bucket with Lid (Globe Scientific, catalog number: IBB003P )
- EVOSTM FL Imaging System (Thermo Fisher Scientific, catalog number: AMF4300 )
- EVOSTM Light Cube, GFP (Thermo Fisher Scientific, catalog number: AMEP4651 )
- EVOSTM Light Cube, Texas Red (Thermo Fisher Scientific, catalog number: AMEP4655 )
- Biotek SynergyTM 2 Multi-mode Detection Microplate/Plate Reader (BioTek Instruments, catalog number: 7160204 )
Software
- Vector NTI software 11.5 (Thermo Fisher Scientific)
- Gen5 1.10 (BioTek Instruments)
- Excel 14.0 (Microsoft)
- Origin 6.0 (OriginLab)
Procedure
- Preparation of plasmid
- The pIM2 vector harbors a multiple cloning site (MCS) and mCherry flanking an internal ribosome binding site (IRES; Figure 1). This enables one to check transfection efficiency by following mCherry fluorescence (Figure 2). A P2X7 receptor gene (XP_002913164.2) is subcloned in MCS using BamHI and XhoI. (Figure 1)

Figure 1. Vector map of pIM2-P2X7. This figure is created using Vector NTI software (Thermo Fisher Scientific). - Amplify the plasmid using an E.Z.N.A. miniprep kit.
- Measure the concentration of plasmid using Nanodrop (normally around 0.5 μg/μl).
- Store the plasmid in a -20 °C freezer.
- The pIM2 vector harbors a multiple cloning site (MCS) and mCherry flanking an internal ribosome binding site (IRES; Figure 1). This enables one to check transfection efficiency by following mCherry fluorescence (Figure 2). A P2X7 receptor gene (XP_002913164.2) is subcloned in MCS using BamHI and XhoI. (Figure 1)
- HEK293 cell preparation (perform in a biosafety cabinet)
- Store aliquots of low passage HEK293 cells in liquid nitrogen.
- Thaw an aliquot of HEK293 cells in a 37 °C water bath and transfer into 10 ml DMEM-FBS medium in a 10 cm dish. Incubate the cells in a CO2 incubator at 37 °C.
- When the cell density reaches 90-100% confluency, aspirate the medium, rinse with 5 ml DPBS, and dissociate with 1 ml trypsin. Split the cells at 1/10 cell density with DMEM-FBS in a new 10 cm dish. Repeat Steps B2 and B3 for maintaining HEK293 until sued in the YO-PRO-1 uptake assay.
- One day before transfection, split 90-100% confluent HEK cells in a 10 cm dish into a 6-well plate by performing Steps B5-B8.
- Rinse the cells with DPBS and detach them with 1 ml trypsin.
- Resuspend the cells by gently pipetting with 12 ml DMEM-FBS.
- Plate 2 ml of resuspended cells into each well of a 6-well plate.
- Incubate the HEK cells overnight in a CO2 incubator.
- Store aliquots of low passage HEK293 cells in liquid nitrogen.
- Transfection (perform in a biosafety cabinet)
- For each well of the 6-well plate, prepare DNA/transfection reagent mixture in the following procedure.
- Add 2 μg of pIM2 plasmid, 5 μl jetPRIME transfection reagent, and 200 μl jetPRIME buffer in a 1.5 ml Eppendorf tube.
- Mix well by vortexing and incubate for 15 min at room temperature (RT).
- Add the DNA/transfection-reagent mixture into a well of the HEK cells on a 6-well plate.
- Incubate the cells for 3 h at 37 °C in a CO2 incubator (No shaking).
- Aspirate the medium and rinse the cells with 2 ml of DPBS.
- Incubate the cells with 0.5 ml trypsin at RT.
- Mix the cells with 3 ml DMEM-PBS by gently pipetting up and down 3 times.
- Split 100 μl of resuspended cells into each well of a poly-L-lysine coated 96-well plate.
- Incubate the cells overnight at 37 °C in a CO2 incubator.
- For each well of the 6-well plate, prepare DNA/transfection reagent mixture in the following procedure.
- YO-PRO-1 uptake assay
- Check the transfection efficiency by mCherry fluorescence using a fluorescent microscope (Figure 2). More than 70% of the cells should show red fluorescence.
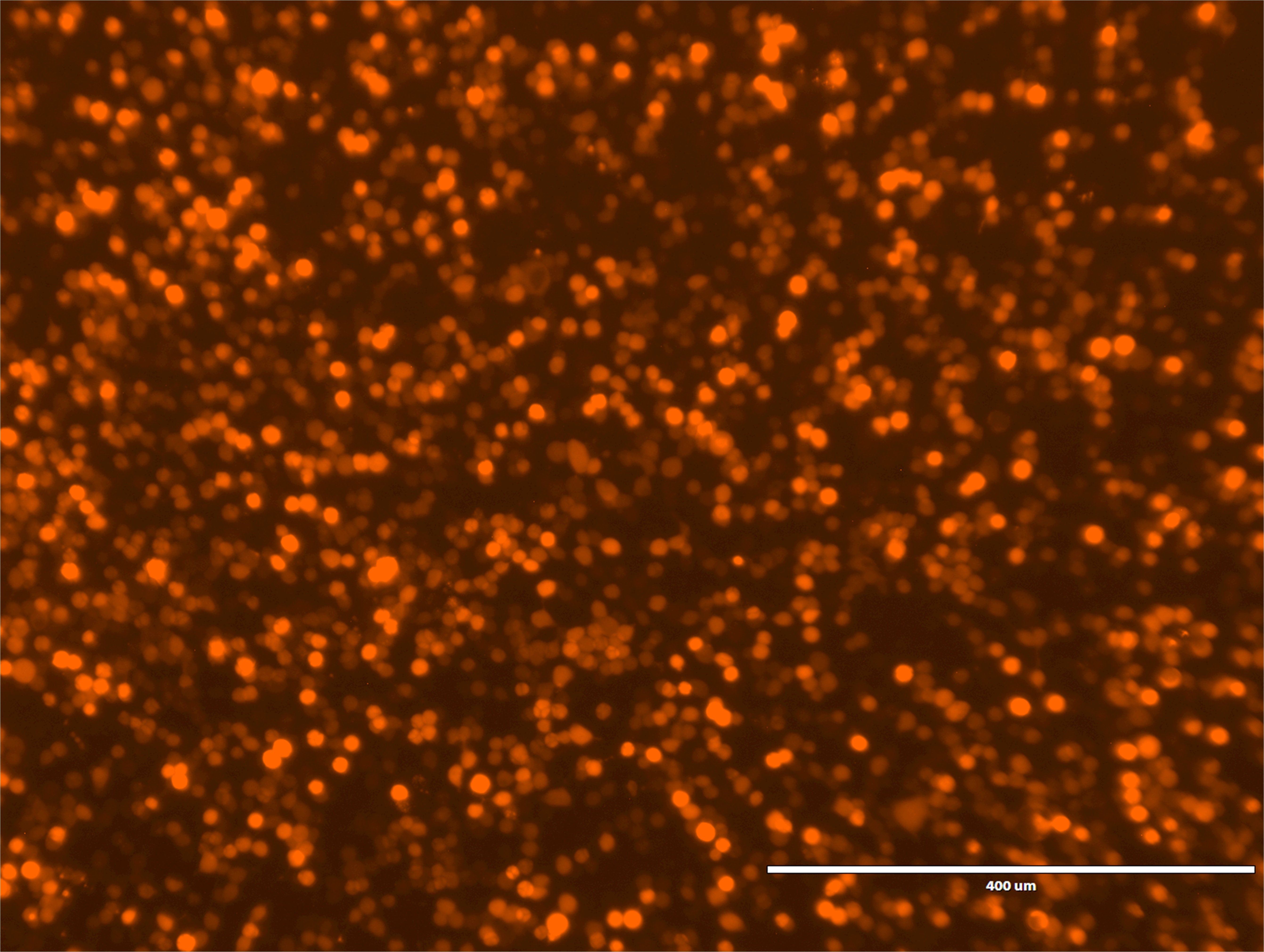
Figure 2. mCherry fluorescence from HEK cells expressing P2X7. Image was obtained using an Evosf1 microscope (10x objective; 10% gain; Ex: 585/29 Em: 624/40). Scale bar = 400 μm. Typically more than 70% of the cells are fluorescent. - Prewarm Assay buffer and YO-PRO-1 solution (10 ml/plate each) at 37 °C in a water bath.
- Replace the DMEM-FBS medium in the 96-well plate with 100 μl/well Assay buffer.
- Aspirate Assay buffer and gently add 100 μl YO-PRO-1 solution in each well. When drugs are tested, include the desired concentration of compounds in the YO-PRO-1 solution.
- Incubate the plate for 15 min at 37 °C in a CO2 incubator.
- Prewarm the plate reader at 37 °C.
- Prepare 11 mM ATP in Assay buffer from 100 mM stock and dispense 100 μl/well in a 96-well plate (no poly-L-lysine coating plastic plate).
- Open Gen5 software on Biotek Synergy 2 Multi-mode Detection Microplate/Plate Reader.
- Click the procedure and create the program as shown in Figure 3.
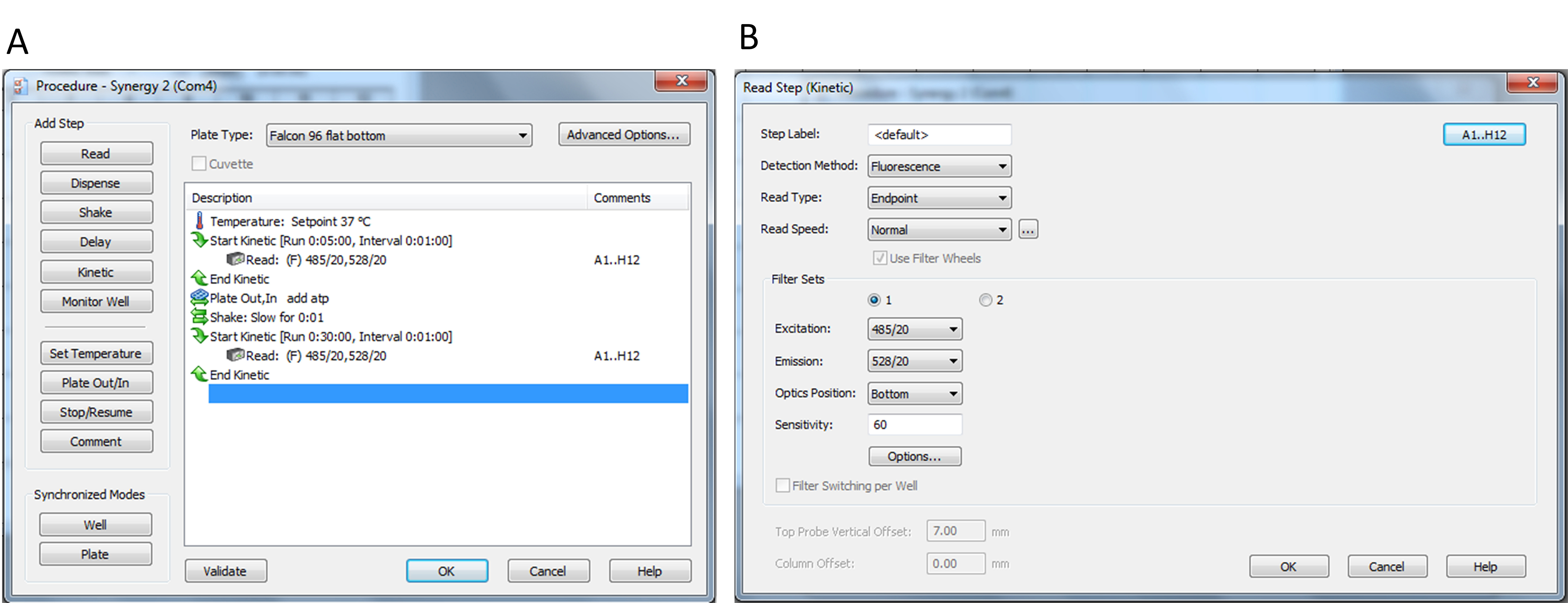
Figure 3. Screenshots of the YO-PRO-1 uptake assay program on Gen5 software. A. Program used in this protocol; B. Input setup for fluorescence measurements. - Select the wells that include samples by clicking the upper right button in Figure 3B.
- Hit "Read" bottom and the tray will come out automatically.
- Insert the 96-well plate on the tray and start the program.
- After measuring the background fluorescence for 5 min, the tray will come out.
- Immediately add 10 μl of 11 mM ATP into each well using a multichannel pipette.
- Push the tray back into the machine and the program continues to measure fluorescence.
- Once the program is finished, click the well in the matrix and export the data (time and fluorescence values) as a Microsoft Excel file.
- Check the YO-PRO-1 fluorescence from the cells under an Evosf1 microscope. If P2X7 was activated by ATP, green fluorescence should be observed (Figure 4B).

Figure 4. YO-PRO-1 fluorescence from HEK cells. Images were obtained using an Evosf1 microscope (10x objective; 10% gain; Ex: 470/22 Em: 525/50). Scale bars = 400 μm. A. HEK cells transfected with Empty pIM2 vector; B. HEK cells transfected with pIM2-P2X7.
- Check the transfection efficiency by mCherry fluorescence using a fluorescent microscope (Figure 2). More than 70% of the cells should show red fluorescence.
Data analysis
- Export the raw data as a Microsoft Excel file.
- Normalized the fluorescence by subtracting the background fluorescence.
- Plot the fluorescence value against the time (Figure 5A).
- Obtain the initial rates of YO-PRO-1 uptake by linear regression analysis on Excel (Figure 5B). Normally, the first 4-6 points are within the linear range.
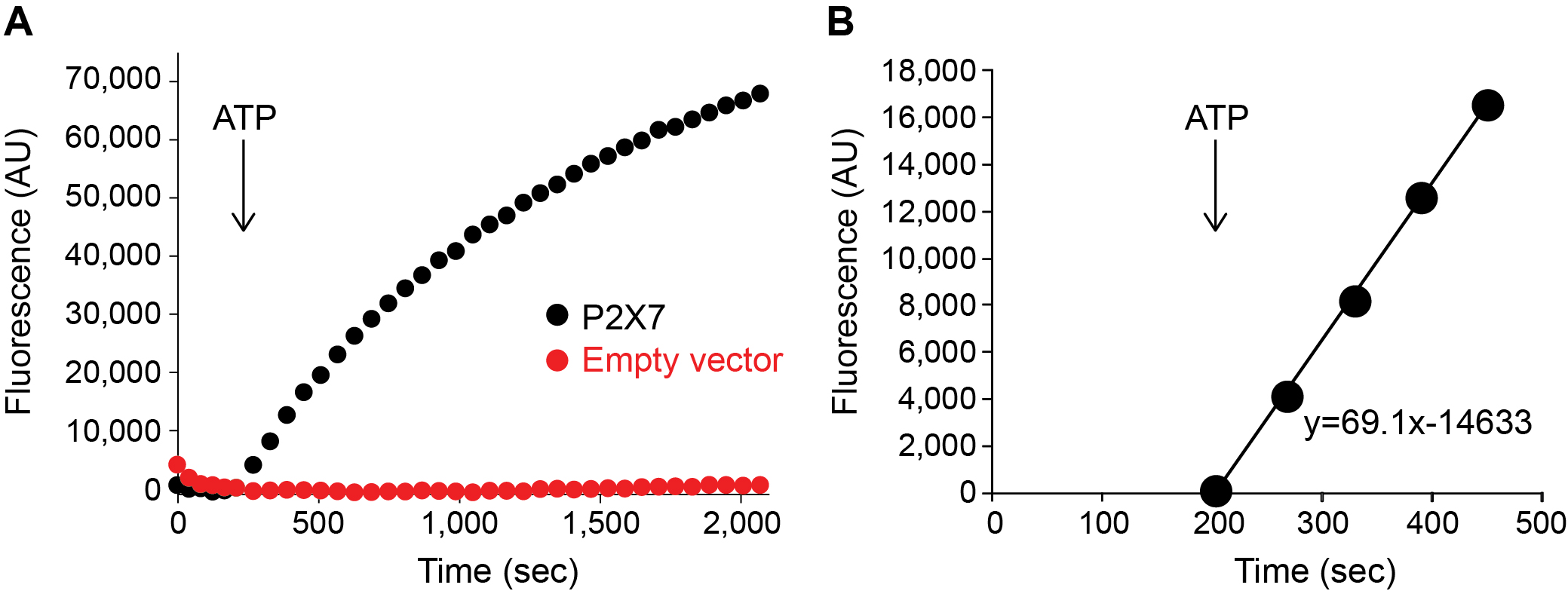
Figure 5. YO-PRO-1 uptake by P2X7 activation. A. P2X7-mediated YO-PRO-1 uptake triggered by 1 mM ATP (black). Vector control (red) shows little accumulation of YO-PRO-1. B. Zoomed in window highlighting the initial linear range of Yo-Pro-1 uptake activity. Slope of this equation (69.10) represents the initial rate for this assay. Graphs were made by Origin software for representation purpose in this figure. - For the measurement of the efficacy of a drug, carry out the YO-PRO-1 uptake assay in the presence of different concentrations of the drug (Figure 6A).
- Plot the initial rates against drug concentrations (Figure 6B).
- Calculate the IC50 value by fitting a Hill equation.
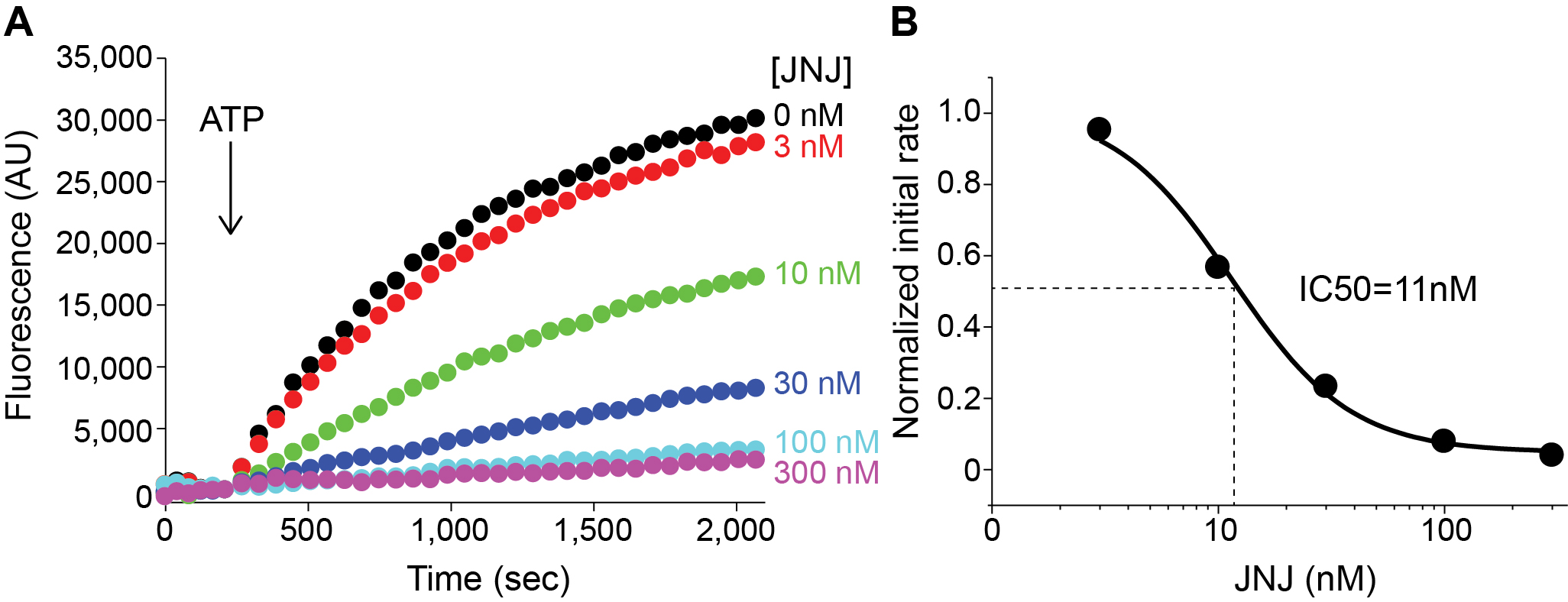
Figure 6. Efficacy of P2X7 specific inhibitor JNJ. A. P2X7-mediated YO-PRO-1 uptake triggered by 1 mM ATP in the absence (Black) or in the presence of JNJ at different concentrations (Color). B. Initial rates of YO-PRO-1 uptake at different concentrations were plotted against JNJ concentrations. These points were fit to a Hill equation using Origin software. - Repeat the assay with HEK cells transfected on different days. Normally 5 different assays are used for quantifying P2X7 activity.
- Calculate the IC50 values from each assay and obtain the average values.
Notes
- Overgrown or unhealthy HEK cells present substantially lower transfection efficiency. In addition, unhealthy cells tend to give higher background of YO-PRO-1 fluorescence as this dye can get into these cells without P2X7 activation.
- Poly-L-lysine coated plate is essential as HEK cells easily come off from the non-coated plate during the washing step.
- YO-PRO-1 is light sensitive. We found fluorescence intensity of YO-PRO-1 significantly went down after a few hours. YO-PRO-1 solution should be prepared just before the fluorescence measurement.
Recipes
Note: Prepare Recipe 1 DMEM-FBS in a tissue culture hood. Other solutions do not need to be sterilized.
- DMEM-FBS
For 500 ml DMEM-FBS, replace 50 ml of DMEM with 50 ml FBS and 500 μl of Gentamicin - Assay buffer
Weigh out the following chemicals in a 500 ml beaker:NaCl 4.3 g HEPES 1.2 g Glucose 1.17 g KCl 74.6 mg CaCl2 7.35 mg - Dissolve in Milli-Q water by mixing with stir bar
- Adjust the pH to 7.4 with NaOH
- Make up the volume to 500 ml with Milli-Q water
- Store the solution in a 4 °C fridge
- Dissolve in Milli-Q water by mixing with stir bar
- YO-PRO-1 Solution (prepare freshly just before the assay)
- Thaw 1 mM YO-PRO-1 stock (provided in DMSO) at RT (stored in a -20 °C freezer)
- Take 55 μl of YO-PRO-1 stock solution into a 15 ml tube and add 11 ml of Assay buffer to make 5 μM YO-PRO-1 solution
- Thaw 1 mM YO-PRO-1 stock (provided in DMSO) at RT (stored in a -20 °C freezer)
- 100 mM ATP stock
- Weigh out 1.1 g of ATP in a 50 ml tube
- Dissolve the ATP in Assay buffer
- Adjust the pH to 7.4
- Make up the volume to 20 ml with Milli-Q water
- Aliquot it into a 1.5 ml tube and store in a -20 °C freezer
- Weigh out 1.1 g of ATP in a 50 ml tube
- 100 mM Inhibitor stock (JNJ)
- Weigh out 9.77 mg JNJ 47965567 in a 1.5 ml tube
- Dissolve it with 200 μl DMSO
- Aliquot it into 1.5 ml tube and store in a -20 °C freezer
- Weigh out 9.77 mg JNJ 47965567 in a 1.5 ml tube
Acknowledgments
This protocol was adapted from our previous work (Karasawa and Kawate, 2016). This work was supported by the National Institutes of Health (GM114379 and NS072869). The authors declare no conflicts of interest.
References
- Alves, L. A., de Melo Reis, R. A., de Souza, C. A., de Freitas, M. S., Teixeira, P. C., Neto Moreira Ferreira, D. and Xavier, R. F. (2014). The P2X7 receptor: shifting from a low- to a high-conductance channel - an enigmatic phenomenon? Biochim Biophys Acta 1838(10): 2578-2587.
- Cankurtaran-Sayar, S., Sayar, K. and Ugur, M. (2009). P2X7 receptor activates multiple selective dye-permeation pathways in RAW 264.7 and human embryonic kidney 293 cells. Mol Pharmacol 76(6): 1323-1332.
- Karasawa, A. and Kawate, T. (2016). Structural basis for subtype-specific inhibition of the P2X7 receptor. Elife 5: e22153.
- Karasawa, A., Michalski, K., Mikhelzon, P. and Kawate, T. (2017). The P2X7 receptor forms a dye-permeable pore independent of its intracellular domain but dependent on membrane lipid composition. Elife 6: e31186.
- Qu, Y., Misaghi, S., Newton, K., Gilmour, L. L., Louie, S., Cupp, J. E., Dubyak, G. R., Hackos, D. and Dixit, V. M. (2011). Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J Immunol 186(11): 6553-6561.
- Rassendren, F., Buell, G. N., Virginio, C., Collo, G., North, R. A. and Surprenant, A. (1997). The permeabilizing ATP receptor, P2X7. Cloning and expression of a human cDNA. J Biol Chem 272(9): 5482-5486.
- Sluyter, R. (2017). The P2X7 Receptor. Adv Exp Med Biol 1051: 17-53.
- Surprenant, A., Rassendren, F., Kawashima, E., North, R. A. and Buell, G. (1996). The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7). Science 272(5262): 735-738.
Karasawa and Kawate. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
Category
Cell Biology > Cell-based analysis > Transport
Molecular Biology > Protein > Ion channel signaling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link


