Advanced Search
Isolation of Phages Infecting Marinomonas mediterranea by an Enrichment Protocol
Published: Jul 5, 2018 DOI: 10.21769/BioProtoc.2921 Views: 6925
Abstract
This protocol describes the isolation of lytic phages infecting the marine bacterium Marinomonas mediterranea from samples of seawater, sand, and seagrass from Posidonia oceanica meadows. It includes the collection of samples, the enrichment method and the isolation and purification of the phages using double layer agar plates. Although the method has been optimized for M. mediterranea, it might be used in the isolation of phages infecting other Marinomonas species and marine bacteria.
Keywords: Marinomonas mediterraneaBackground
CRISPR-Cas systems provide adaptive immunity against genetic infection in prokaryotes by acquiring short segments of nucleic acids of the invasive element (spacers). These “molecular memories of infection” are used to produce guide RNAs that target Cas nucleases to the pathogen when a new infection occurs. The spacers are stored directly in the genome of the host, interspersed between directed repeats of the CRISPR arrays. The analysis of the repertoire of spacer sequences in prokaryotic genomes as well as in metagenomic datasets has highlighted a vast diversity of undiscovered genetic pathogens that are the targets of CRISPR-Cas systems (Shmakov et al., 2017). The isolation of phages infecting microorganisms of interest can provide important insights into the mechanism of action of the CRISPR-Cas systems in a natural context.
In this work, we provide an easy and inexpensive method for isolating phages infecting the model bacterium Marinomonas mediterranea, from the natural environment of this microorganism. This bacterium has two different CRISPR-Cas systems: a canonical type I-F system, and also one type III-B system capable of acquiring spacers not only from DNA but also from RNA (Silas et al., 2016). This protocol could be easily adapted for the isolation of other phages from marine environments, which contain an enormous diversity of uncharacterized viruses.
Materials and Reagents
- Cotton gauze swabs 20 x 20 cm (Gaspunt®, catalog number: PI132001 )
- Cellulose esters filters 0.45 µm, 47 mm diameter (Merck, MF-MilliporeTM, catalog number: HAWP04700 )
- 90 mm Petri Dishes (Thermo Fisher Scientific, Thermo ScientificTM, SterilinTM, catalog number: 101VR20 )
- 96-Well Microplates (Thermo Fisher Scientific, Thermo ScientificTM, NuncTM MicroWellTM, catalog number: 269620 ) for OD measurements in the spectrophotometer
- Disposable culture borosilicate glass tubes 16 x 100 mm (Corning, PYREX®, catalog number: 99445-16 )
- Syringe Luer-LokTM 10 ml (BD, catalog number: 309604 )
- Syringe filters, PES, 0.2 µm, 33 mm diameter, sterile (Fisher Scientific, FisherbrandTM, catalog number: 15206869 )
- Sea Salts (Sigma-Aldrich, catalog number: S9883 )
- Marine Agar (Conda, catalog number: 1059 )
- Glycerol (Fisher Scientific, Fisher BioReagentsTM, catalog number: BP229-1 )
- Sodium Chloride (NaCl) (Scharlab, catalog number: SO02271000 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Scharlab, catalog number: MA00851000 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Merck, EMSURE®, catalog number: 1.05833.1000 )
- Potassium chloride (KCl) (Merck, EMSURE®, catalog number: 1.04936.0500 )
- Calcium chloride 2-hydrate (CaCl2·2H2O) (ITW Reagents Division, PanReac AppliChem, catalog number: 131232.1211 )
- Sodium hydroxide (NaOH) (ITW Reagents Division, PanReac AppliChem, catalog number: 141687 )
- European bacteriological agar (Conda, catalog number: 1800 )
- Glucose (VWR, NORMAPUR®, catalog number: 24370.294 )
- Iron (III) citrate hydrate (Merck, catalog number: 1.03862 )
- Tri-sodium citrate 2-hydrate (ITW Reagents Division, PanReac AppliChem, catalog number: 141655 )
- Di-potassium hydrogen phosphate (VWR, RECTAPUR®, catalog number: 26930.293 )
- Bacteriological peptone (Conda, catalog number: 1616 )
- Yeast extract (Conda, catalog number: 1702 )
- MMC2G medium (see Recipes)
Equipment
- NalgeneTM 250 ml Polycarbonate Centrifuge Bottles with Sealing Closure (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3140-0250 )
- 47 mm Glass filtration set with funnel, fritted membrane support, clamp, silicone rubber stopper and receiver kitasato flask
- Pipettes (P1000, P100, P10)
- 100 ml, 1 L and 2 L glass flasks
- Flask shaker (GFL-Gesellschaft für Labortechnik, model: 3005 )
- Centrifuge for high volume samples (Thermo Fisher Scientific, model: SorvallTM RC 5B plus with GSA rotor)
- Centrifuge for low volume samples (Eppendorf, model: 5430R )
- Vacuum pump (KNF, model: N 86 KN.18 )
- 25 °C incubator (Kgroup, Koxka, model: API-6/1 MIL )
- Microplate SpectroPhotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: Multiskan® Spectrum )
- Water bath (LAUDA-Brinkmann, model: Aqualine AL25, catalog number: L000612 )
- pH meter (HACH, Crison, model: Basic 20 )
- Dri-block® Dry Thermoblock (Cole-Parmer, Techne, model: DB-2D )
Procedure
- Sample collection
Marinomonas mediterranea forms part of the microbiota associated with the seagrass Posidonia oceanica (Espinosa et al., 2010). Take samples of seawater and sand from the surroundings of this seagrass, as well as samples from different parts of the Posidonia oceanica plant itself in glass bottles. Keep plant and sand samples in seawater. Keep the samples at 4 °C until processed. We typically processed the samples within 24-48 h.
Note: In various attempts to isolate phages infecting Marinomonas mediterranea, we have obtained plaques only when using the sand and seawater samples, never from the plant samples. - Processing of the samples for the enrichment protocol
The processing protocol is dependent on the type of sample.- For seawater samples
- Filter the water samples through 7-8 layers of unfolded cotton gauze swabs placed on a glass funnel under non-sterile conditions.
- Centrifuge the filtrate at 5,000 x g, 10 min, 4 °C.
- Filter the supernatant through 0.45 µm Millipore HA filters placed on a fritted glass base using a 47 mm glass filtration set connected to a vacuum pump.
- Keep the filtrate in glass flasks at 4 °C until the enrichment protocol is performed (Procedure C below). We have obtained plaques from filtrates that had been stored up to 11 days at 4 °C.
- Filter the water samples through 7-8 layers of unfolded cotton gauze swabs placed on a glass funnel under non-sterile conditions.
- For plant samples, suspend the plant tissue at 20% (w/v) in seawater (or sterile SeaSalts solution), mash it with a hand-held kitchen blender, filter the samples through cotton gauze swabs, and follow the rest of steps as with the water samples.
- For sand samples, add wet sand to untreated seawater at 20% in a flask just before beginning the enrichment (Procedure C). No further filtration is necessary for this sample.
Note: The sand concentration is approximate, as it is not dried before weighing.
- For seawater samples
- Enrichment of phages infecting M. mediterranea
Use M. mediterranea strains that might enhance the probabilities of phage propagation as hosts for the phage enrichments. We have succeeded using either a mutant with a deletion of the III-B CRISPR-Cas system (∆III-B mutant strain) or a regulatory mutant, T103, which shows defects in CRISPR-Cas expression (Silas et al., 2017).- Day 1
Streak the M. mediterranea host strain on a marine agar plate from a -80 °C frozen glycerol stock and incubate for 48 h at 25 °C. We keep the plate inside a cardboard box for the first overnight incubation, but place it inside a plastic bag until the second day so that the medium does not get very dry. After they are grown, the plates are kept at 15 °C. Do not keep the plates at 4 °C since this adversely affects cell viability. - Day 3
Pick 2-3 isolated colonies and inoculate into 10 ml MMC2G broth in a 100 ml flask at 25 °C overnight at 130 rpm.
Note: We never fill in excess of 1/5th of the total volume of the flask to ensure good aeration of the culture as M. mediterranea is a strict aerobe. - Day 4
- Measure the OD600 of the overnight culture in a microplate spectrophotometer using a 10-1 dilution of the culture added to a 96-well microplate. This culture is used to inoculate a new 100 ml MMC2G culture in a 1 L flask such that the initial OD600 is 0.05. Grow at 25 °C (130 rpm) until the culture reaches late exponential phase (OD600 of 0.6, approximately 4.5 h of incubation).
- Add 20 ml of the late exponential phase culture to every 180 ml of the processed environmental sample (from Procedure B: seawater, seagrass or sand) supplemented with 0.5% peptone and 0.1% yeast extract (we add 9 ml peptone and 1.8 ml yeast extract from sterile-filtered 10% stocks). Incubate this enrichment culture in a 1 L volume glass flask at 75 rpm, 25 °C, 16 h (Figure 1).
- Prepare another 100 ml flask (labeled control 1) with 18 ml of the processed environmental sample (from Procedure B) containing 0.5% peptone and 0.1% yeast extract, and add 2 ml of MMC2G (but no bacteria). Incubate it under the same conditions, 75 rpm, 25 °C, 16 h, as the enrichment assay (Figure 1).
- Pick 2-3 isolated colonies of the host strain from the plate from Day 1 and inoculate 10 ml MMC2G broth in a 100 ml flask at 25 °C overnight at 130 rpm.
- Measure the OD600 of the overnight culture in a microplate spectrophotometer using a 10-1 dilution of the culture added to a 96-well microplate. This culture is used to inoculate a new 100 ml MMC2G culture in a 1 L flask such that the initial OD600 is 0.05. Grow at 25 °C (130 rpm) until the culture reaches late exponential phase (OD600 of 0.6, approximately 4.5 h of incubation).
- Day 5
- Measure the OD600 of the overnight culture of the host strain and use this to inoculate a new 100 ml MMC2G culture in a 1 L flask such that the initial OD600 is 0.05. Grow at 25 °C (130 rpm) until the culture reaches late exponential phase (OD600 of 0.6).
- Prepare 5 replicate borosilicate glass tubes for each enrichment (Step C3b) and 3 replicates for the control 1 flask (Step C3c), containing 3.6 ml of fresh MMC2G medium and 0.4 ml of the late exponential M. mediterranea culture from Step C4a (Figure 1).
- Add 1 ml of each enrichment or control 1 culture, filtered through 0.2 µm polystyrene sulfonate (PSS) sterile filters, into each of corresponding replicate tubes (Figure 1).
- Set another 3 replicate tubes named control 2 that won’t have any 1 ml enrichment filtrate from Day 4, but 1 ml additional fresh MMC2G medium instead (Figure 1).
- Incubate all the tubes without shaking at 25 °C overnight.
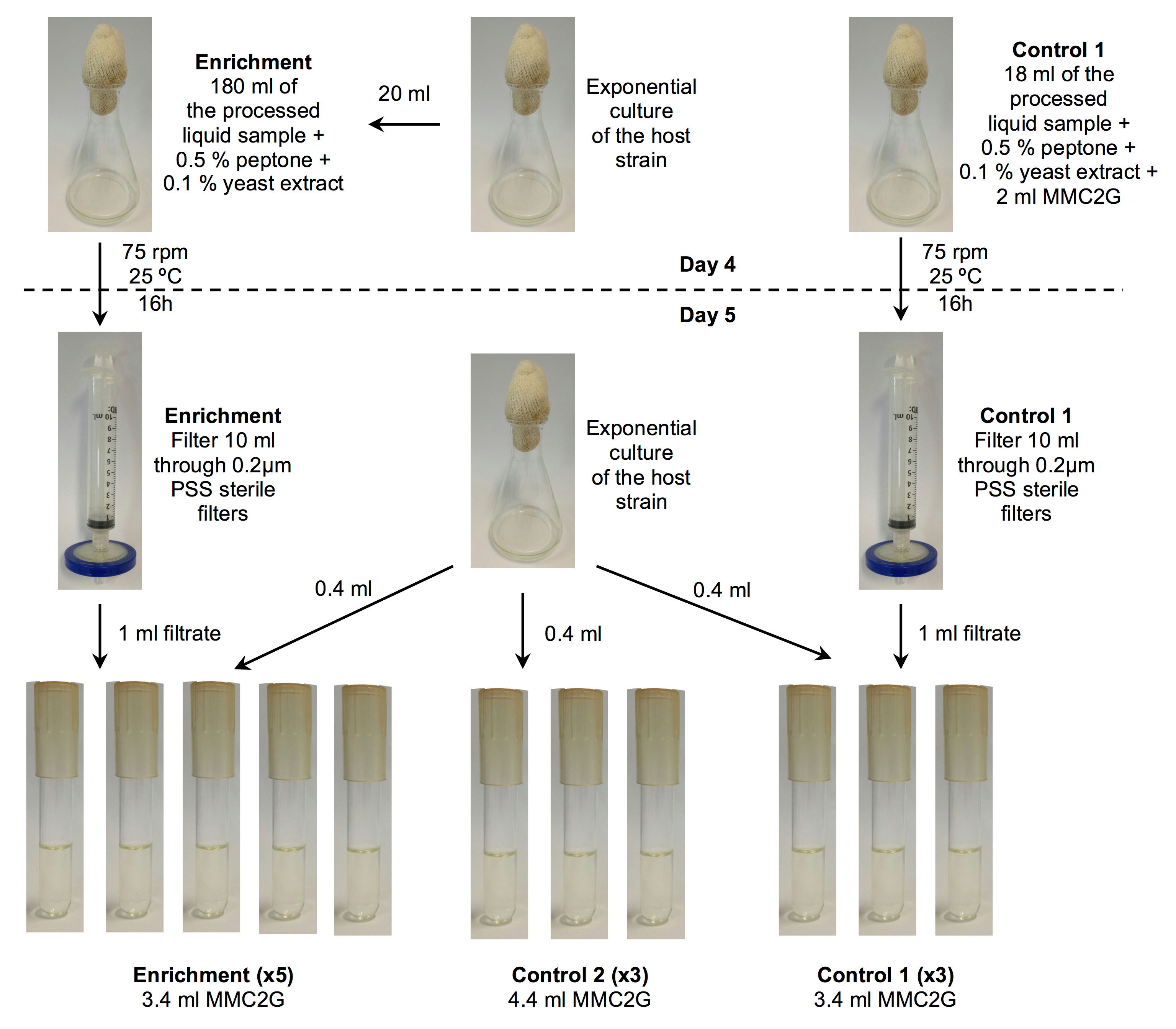
Figure 1. Schematics of the procedure for the days 4 and 5 of the enrichment protocol (Step C). The enrichment and control 1 are set up on Day 4 from the processed environmental samples and an exponential phase culture of the host. The enrichment continues on the Day 5 by inoculating tubes with the overnight enrichment and control 1 cultures. Control 2 is set up on Day 5 using only the exponential phase host culture, but no processed environmental samples. - Measure the OD600 of the overnight culture of the host strain and use this to inoculate a new 100 ml MMC2G culture in a 1 L flask such that the initial OD600 is 0.05. Grow at 25 °C (130 rpm) until the culture reaches late exponential phase (OD600 of 0.6).
- Day 6
- Follow the OD600 of the replicate tubes making a measurement approximately every 12 h. It may take more than one day of incubation to observe a decrease in the optical density of some of the enrichment tubes compared to the control 2 tubes. We consider an enrichment tube as positive if it has a decrease in the OD600 ≥ 50% compared to control 2.
- A decrease in the OD600 of any of the tubes from control 1 may be due to growth inhibitors such as toxic compounds or lytic microorganisms (such as bdellovibrios), which may be interfering in the enrichment cultures as well.
- In the case of a positive result in the enrichment tubes, keep 1 ml at 4 °C and filter the rest of that sample through 0.2 µm PSS sterile filters. Use this filtrate to test for the presence of phages in the samples by the plaque assay (Procedure D, below). Some phages could be retained by the filters, so in case no plaques are obtained, it is possible to go back and test the 1 ml unfiltered aliquot stored at 4 °C.
- Follow the OD600 of the replicate tubes making a measurement approximately every 12 h. It may take more than one day of incubation to observe a decrease in the optical density of some of the enrichment tubes compared to the control 2 tubes. We consider an enrichment tube as positive if it has a decrease in the OD600 ≥ 50% compared to control 2.
- Day 1
- Check for the presence of phages in the enrichments by double layer assays
To perform a double layer plaque assay, a phage aliquot is incubated with susceptible cells to allow phages to attach to cells. Next, this mixture is poured together with melted agar medium (the upper layer) on plates already containing a layer of solid sterile medium (the bottom layer). When the plates are incubated, the infected cells release phage progeny. The spread of the new phages is restricted to neighboring cells by diffusion through the solid medium. Therefore, each infectious particle produces a clear circular zone of lysed cells called a plaque, visible to the naked eye.- Place the plates for the double layer plaque assay (containing the bottom layer of MMC2G medium) at room temperature.
- Dilute an overnight culture of the strain to be tested (for example, M. mediterranea MMB-1 with the CRISPR-Cas loci deleted) with fresh medium to an OD600 of 0.12. In our hands an overnight culture inoculated the previous afternoon works well, as does an exponential phase culture prepared that morning.
- Mix 0.9 ml of the diluted culture with 0.1 ml of the phage filtrate from Step C5c in an Eppendorf tube. Depending on the phage concentration, it may be necessary to use 10-fold dilutions of the phage suspension to get plaques instead of a fully lysed plate. In this case, use the MMCbase (see Recipes) to make the phage dilutions.
- Incubate the mixture of phages and bacteria without shaking at room temperature for about 30 min and then add it to a tube containing the melted top agar kept at 45 °C in a dry thermoblock. Mix rapidly (but avoiding the formation of bubbles by placing the tube vertically between the palm of both hands and rolling forward and backward about 5 times) and distribute it evenly on top of the bottom layer. Let the plates sit until the top agar solidifies (30 min-1 h). Invert the plates and incubate them at 25 °C overnight.
- The presence of phages is revealed by the appearance of plaques in the lawn of host cells (Figure 2).
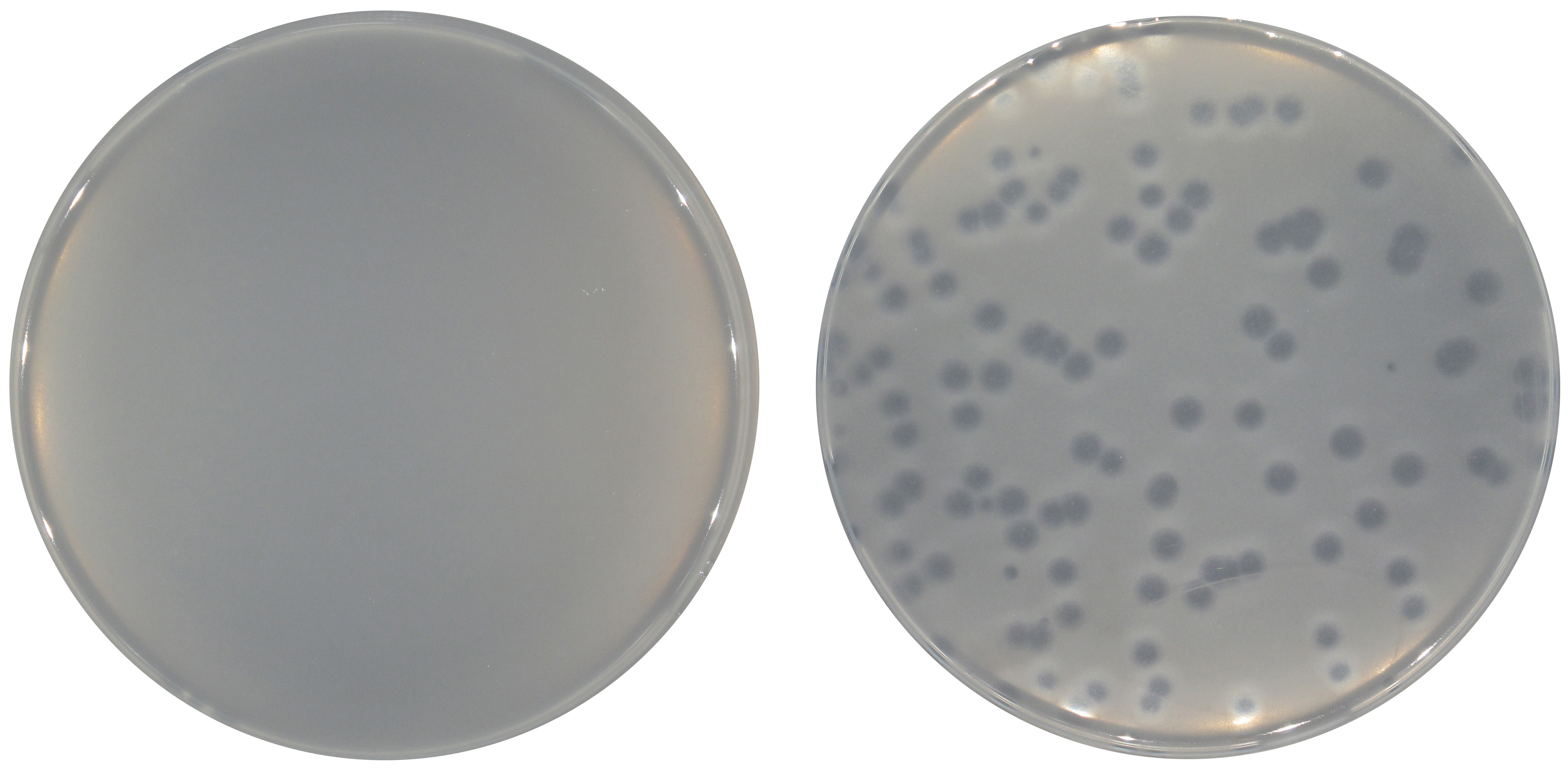
Figure 2. Double layer assay with the appearance of phage plaques. Phage CPG1g plaques on a lawn of M. mediterranea MMB-1 ∆III-B mutant strain (right) compared to a control lawn without any phage addition (left).
- Place the plates for the double layer plaque assay (containing the bottom layer of MMC2G medium) at room temperature.
- Isolation of the virus by plaque purification
- Choose a plate with only a few well-separated plaques and retrieve a sample from a single plaque by picking a bit of the top agar layer using a 200 µl sterilized pipette tip held perpendicularly to the surface of the plate. Choose a plaque far from others so that the sample is not contaminated with other phages diffusing through the agar. Avoid touching the bacteria surrounding the plaque.
- Place the tip in 1 ml of MMC2G broth and gently pipette up and down a few times to release the phage. Discard the tip. Use this broth for another dilution series and plaque assay using an exponential phase culture of the host. Once plaques are obtained, repeat the isolation procedure from a single plaque to be sure that only a single clone of the virus has been selected.
- Choose a plate with only a few well-separated plaques and retrieve a sample from a single plaque by picking a bit of the top agar layer using a 200 µl sterilized pipette tip held perpendicularly to the surface of the plate. Choose a plaque far from others so that the sample is not contaminated with other phages diffusing through the agar. Avoid touching the bacteria surrounding the plaque.
- Preparation of a virus working solution and storage of the isolated virus
- Day 1
To store a phage that was isolated from a plaque, add 4.35 ml of fresh MMC2G or Marine broth, 400 μl of a culture of the host at exponential phase of growth and 10 μl of the suspension with phages from the plaque to a glass tube. Incubate the tube in static conditions at 25 °C overnight. Also set up a control tube without the plaque suspension to compare optical densities the next day. - Day 2
- The tube containing the phage and bacteria should be lysed and should have lower turbidity than the control.
- Filter the contents of that tube through 0.2 µm syringe filters in order to eliminate any remaining bacteria in the sample.
- Mix 800 μl of the filtrate with 200 μl of sterile glycerol, mix by pipetting and immediately store the stock at -80 °C.
- To recover phage from the stock, scrape off a bit of the frozen mass with a sterile tip and introduce the whole tip in a tube containing fresh medium and an exponential phase growth of the host (as described in Step F1).
- The tube containing the phage and bacteria should be lysed and should have lower turbidity than the control.
- Day 1
Recipes
- MMC2G medium
Note: This is a complex medium prepared with MMCbase supplemented with citrate, phosphate and glucose solutions after autoclaving.- Prepare the MMCbase by dissolving in 800 ml of distilled water (in a 2 L flask) the following salts:
NaCl 20.00 g
MgSO4·7H2O 7.00 g
MgCl2·6H2O 5.30 g
KCl 0.70 g
CaCl2·2H2O 1.25 g
Peptone 5.00 g
Yeast Extract 1.00 g - Adjust the pH to 7.4 using a NaOH solution
- Bring the total volume to 1 L with distilled water
- As a stock for MMC2G liquid medium, add the MMCbase to a bottle that will be stored, after autoclaving, at room temperature. For solid medium, add 15 g of European Bacteriological Agar (this yields 1.5% agar plates). For the double layer assays, add 0.8% agar for the bottom layer, which will be poured onto plates. For the top layer prepare the medium with 0.6% agar in small bottles
- Autoclave the MMCbase
- Finalization of the medium for its usage
- When we want to prepare MMC2G liquid medium, we have the MMCbase liquid in a bottle at room temperature and we add the following citrate, phosphate and glucose solutions from the stocks immediately before the inoculation. These components must be autoclaved separately, and the stocks are kept at room temperature in the lab and can be used every time new media are prepared:1)Hydrated ferric citrate (III) at 1% plus sodium citrate at 9%. This solution is added at a 1/1,000 dilution (10 μl/10 ml of medium)2)K2HPO4 1 M: Add 4 μl/10 ml of medium to get a final 0.4 mM concentration3)Glucose 20%: 100 μl/10 ml of medium to get a final 0.2% concentration
- To prepare MMC2G plates, after sterilization keep the MMCbase agar at 50 °C in a water bath and add the citrate solution, phosphate and glucose just before pouring the plates. Once the plates are solid, keep the medium at 15 °C until it is going to be used
- To prepare the top layer, on the day that you are making the double layers, melt the top agar that was prepared in a small bottle, by placing it in boiling water. After it is melted, put in a water bath at 50 °C. Add the citrate, phosphate and glucose from the stock solutions and distribute it in tubes with 3.5 ml of media. Keep it melted at 45 °C until you make the double layers
- When we want to prepare MMC2G liquid medium, we have the MMCbase liquid in a bottle at room temperature and we add the following citrate, phosphate and glucose solutions from the stocks immediately before the inoculation. These components must be autoclaved separately, and the stocks are kept at room temperature in the lab and can be used every time new media are prepared:
- Prepare the MMCbase by dissolving in 800 ml of distilled water (in a 2 L flask) the following salts:
Acknowledgments
This work has been supported by the grant BFU2017-85464-P (Ministerio de Economía, Industria y Competitividad, Spain). This protocol has been adapted from an “Amplification method” previously described by Suttle (1993). We are thankful to JA García Charton for continuously providing us with the marine samples from the Mediterranean Sea. The authors declare that there are no conflicts of interest.
References
- Espinosa, E., Marco-Noales, E., Gomez, D., Lucas-Elio, P., Ordax, M., Garcias-Bonet, N., Duarte, C. M. and Sanchez-Amat, A. (2010). Taxonomic study of Marinomonas strains isolated from the seagrass Posidonia oceanica, with descriptions of Marinomonas balearica sp. nov. and Marinomonas pollencensis sp. nov. Int J Syst Evol Microbiol 60(Pt 1): 93-98.
- Shmakov, S. A., Sitnik, V., Makarova, K. S., Wolf, Y. I., Severinov, K. V. and Koonin, E. V. (2017). The CRISPR spacer space is dominated by sequences from species-specific mobilomes. MBio 8(5): e01397-17.
- Silas, S., Lucas-Elio, P., Jackson, S. A., Aroca-Crevillen, A., Hansen, L. L., Fineran, P. C., Fire, A. Z. and Sanchez-Amat, A. (2017). Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. Elife 6: e27601.
- Silas, S., Mohr, G., Sidote, D. J., Markham, L. M., Sanchez-Amat, A., Bhaya, D., Lambowitz, A. M. and Fire, A. Z. (2016). Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351(6276): aad4234.
- Suttle, C. A. (1993). Chap 15: Enumeration and isolation of viruses. In: Kemp, P. F., Sherr, B. F., Sherr, E. B. and Cole, J. J. (Eds). Handbook of Methods in Aquatic Microbial Ecology. Lewis Publ pp: 121-134.
Lucas-Elio et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
Category
Microbiology > Microbial physiology > Interspecific competition
Cell Biology > Cell isolation and culture > Virus isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link



