- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bimolecular Fluorescence Complementation (BiFC) Assay for Direct Visualization of Protein-Protein Interaction in vivo
Published: Vol 3, Iss 20, Oct 20, 2013 DOI: 10.21769/BioProtoc.935 Views: 24666
Reviewed by: Lin FangFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Urbi Mukhopadhyay [...] Sagar Bhogaraju
Jun 20, 2025 2435 Views

Quantifying Intracellular Distributions of HaloTag-Labeled Proteins With SDS-PAGE and Epifluorescence Microscopy
Julia Shangguan and Ronald S. Rock
Jul 20, 2025 2501 Views

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1323 Views
Abstract
Bimolecular Fluorescence Complementation (BiFC) assay is a method used to directly visualize protein-protein interaction in vivo using live-cell imaging or fixed cells. This protocol described here is based on our recent paper describing the functional association of human chromatin adaptor and transcription cofactor Brd4 with p53 tumor suppressor protein (Wu et al., 2013). BiFC was first described by Hu et al. (2002) using two non-fluorescent protein fragments of enhanced yellow fluorescent protein (EYFP), which is an Aequorea victoria GFP variant protein, fused respectively to a Rel family protein and a bZIP family transcription factor to investigate interactions between these two family members in living cells. The YFP was later improved by introducing mutations to reduce its sensitivity to pH and chloride ions, thus generating a super-enhanced YFP, named Venus fluorescent protein, without showing diminished fluorescence at 37 °C as typically observed with EYFP (Nagai et al., 2006). The fluorescence signal is regenerated by complementation of two non-fluorescent fragments (e.g., the Venus N-terminal 1-158 amino acid residues, called Venus-N, and its C-terminal 159-239 amino acid residues, named Venus-C; see Figure 1A and Gully et al., 2012; Ding et al., 2006; Kerppola, 2006) that are brought together by interaction between their respective fusion partners (e.g., Venus-N to p53, and Venus-C to the PDID domain of human Brd4; see Figure 1B and 1C). The intensity and cellular location of the regenerated fluorescence signals can be detected by fluorescence microscope. The advantages of the proximity-based BiFC assay are: first, it allows a direct visualization of spatial and temporal interaction between two partner proteins in vivo; second, the fluorescence signal provides a sensitive readout for detecting protein-protein interaction even at a low expression level comparable to that of the endogenous proteins; third, the intensity of the fluorescence signal is proportional to the strength of protein-protein interaction (Morell et al., 2008); and fourth, the BiFC signals are derived from intrinsic protein-protein interaction, rather than from extrinsic fluorophores that may not reflect true protein-protein interaction due to their nonspecific association with cellular macromolecules or subcellular compartments. However, some limitations of BiFC include slow maturation (T1/2 ~ 1 hour) of an eventually stable BiFC complex (Hu et al., 2002), making it unsuitable for real-time observation of transient interaction that disappears prior to BiFC detection, and enhanced BiFC background at high expression levels due to fusion-independent association between two non-fluorescent fragments association. BiFC signals generated by in vivo protein-protein interaction can be validated by amino acid mutation introduced at the protein-protein contact surfaces. This imaging technique has been widely used in different cell types and organisms (Kerppola, 2006).
Keywords: BiFC

Figure 1. Protein fragments of Venus (super enhanced YFP) constructs. A. Venus protein (amino acids 1-239; accession number: CAO79509) was dissected into two fragments at residue 158 to generate Venus N-terminus (top) and Venus C-terminus (bottom). B. Schematic drawing of Venus-N-p53 and Venus-C-PDID fusion fragments. Venus-N-p53 and Venus-C-PDID contain Venus-N-terminus and Venus-C-terminus fused respectively to p53 (amino acids 1-393; Gully et al., 2012) and the phosphorylation-dependent interaction domain (PDID, amino acids 287-530) of human Brd4 (Wu et al., 2013), in which a flexible linker containing two copies of Gly4Ser peptide is introduced to allow optimal space contacts between Venus-N-terminus and Venus-C-terminus and also to prevent steric hindrance between the Venus fragment and its fused protein of interest. AscI and XbaI indicate the positions of restriction enzyme-cutting sites used for generating fusions from PCR-amplified DNA fragments. An initiation codon for methionine (M) was added to allow translation of Venus-C-PDID. It should be noted that, although linker peptides ranging from 5 to 17 amino acids are often used (Remy and Michnick, 2007), the exact length and the sequence nature of the linkers have not been systematically analyzed (Kerppola, 2013). C. BiFC fluorescence signal is produced when Venus-N and Venus-C are in close proximity brought together via p53-PDID interaction in the cell.
Materials and Reagents
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F2442 )
- Antibiotics (Penicillin/Streptomycin) (Sigma-Aldrich, catalog number: P0781 )
- Cell culture medium (Complete: With 10% FBS and antibiotics; Antibiotic-free: with 10% FBS only)
- Formaldehyde (Thermo Fisher Scientific, catalog number: F79-500 )
- Triton X-100 (Sigma-Aldrich, catalog number: 79284 )
- BSA (Sigma-Aldrich, catalog number: A3059 )
- Lipofectamine 2000 (Life Technologies, Invitrogen™, catalog number: 11668-019 )
- Venus-N-p53 (Gully et al., 2012) (Santa Cruz, catalog number: sc8334 ) and Venus-C-PDID (Wu et al., 2013) plasmids (Santa Cruz, catalog number: sc5384 ) (see Figure 1B)
- Primary antibodies against Venus
e.g. Anti-full-length-GFP antibody (Santa Cruz, catalog number: sc8334 or sc9996 )
Anti-C-terminal GFP antibody (Santa Cruz, catalog number: sc5384)
or β-actin (Sigma-Aldrich, catalog number: A5441 )
- Secondary antibody conjugated with a fluorescence dye emitting wavelength other than that of Venus (excitation 488 nm, emission 515 ± 15 nm) or Hoechst 33258 (excitation 350 nm, emission 461 nm), for example, Alexa Fluor® from Life Technologies
- Sodium chloride (Thermo Fisher Scientific, catalog number: 7647-14-5 )
- Potassium chloride (Thermo Fisher Scientific, catalog number: 7447-40-7 )
- Sodium phosphate dibasic heptahydrate (Na2HPO4.7H2O) (Thermo Fisher Scientific, catalog number: S373500 )
- Potassium phosphate monobasic (KH2PO4) (Thermo Fisher Scientific, catalog number: 7778-77-0 )
- Aluminum foil (grocery store)
- Permanent mounting medium (Vector Laboratories, catalog number: H-5000 )
- Microscope slides (Thermo Fisher Scientific, catalog number: 12-544-7 )
- Nail polish (grocery store)
- 10x Phosphate Buffered Saline (PBS) (see Recipes)
- 3.7% Formaldehyde (freshly prepared) (see Recipes)
- Phosphate Buffered Saline with Triton X-100 and BSA (PBSTB) (see Recipes)
- Hoechst 33258 (Sigma-Aldrich, catalog number: 861405 ) (see Recipes)
Equipment
- Glass bottom culture dish (35-mm glass bottom plate containing a 14-mm center microwell, poly-D-lysine coated) (MatTek, catalog number: P35GC-1.5-14-C )
- Tissue culture hood (NuAire, model: Class II, Type A2 )
- 37 °C cell culture incubator (Thermo Fisher Scientific, Forma®, model: Series II , water-jacketed and HEPA filtered)
- Confocal fluorescence microscope (Nikon, model: Eclipse TE-2000E/C1 )
- Rocker (Labnet International, model: Rocker 25 )
Software
- NIS Elements Basic Research (version 2.2)
- Nikon EZ-C1 Free Viewer (version 3.90)
Procedure
Note: Steps 1 to 7 performed in a tissue culture hood; steps 8 to 10 done on regular bench.
- The day before transfection: Seed log-phase growing cells of interest (2 x 105 cells in 2 ml) in a 35-mm glass bottom culture dish and allow overnight incubation for proper cell attachment and expansion in a 37 °C cell culture incubator.
Note: Optimum cell cultures are 30% to 40% confluent with a low percentage of overlapping cells on the day of transfection.
Note: Pre-warm culture medium and 1x PBS to 37 °C.
- Rinse cells twice with 2 ml of 37 °C 1x PBS.
Note: Avoid center glass area when pipetting solutions at all steps.
- Replace with 1 ml antibiotic-free medium.
- Co-transfect Venus-N-p53 and Venus-C-PDID constructs with Lipofectamine 2000 according to manufacture's instructions.
Note: 0.5 μg of each construct (total 1 μg DNA) plus 2.5 μl of Lipofectamine 2000 in 25 μl of Opti-MEM works well with HCT116 cells.
Note: Negative controls, such as Venus-N-p53 with Venus-C linked to a non-interacting protein (or domain, e.g., Brd4 amino acids 149-284 described in Wu et al., 2013), Venus-N-p53 with Venus-C, Venus-N with Venus-C-PDID, or Venus-N with Venus-C, should be included in parallel for comparison.
- Leave cells at 37 °C in a cell culture incubator.
- Replace transfection medium with complete medium 6 hours post-transfection.
- Incubate cells for 24 h at 37 °C in a cell culture incubator.
Note: Incubation time after transfection can vary by the level of protein expression. Pilot experiments to test the optimum expression time and the levels of protein expression for Venus-N-p53 and Venus-C-PDID are beneficial (see Figure 2).
Note: The amounts of transiently expressed proteins should be titrated to the levels of the endogenous proteins, reflecting endogenous protein interaction in vivo.
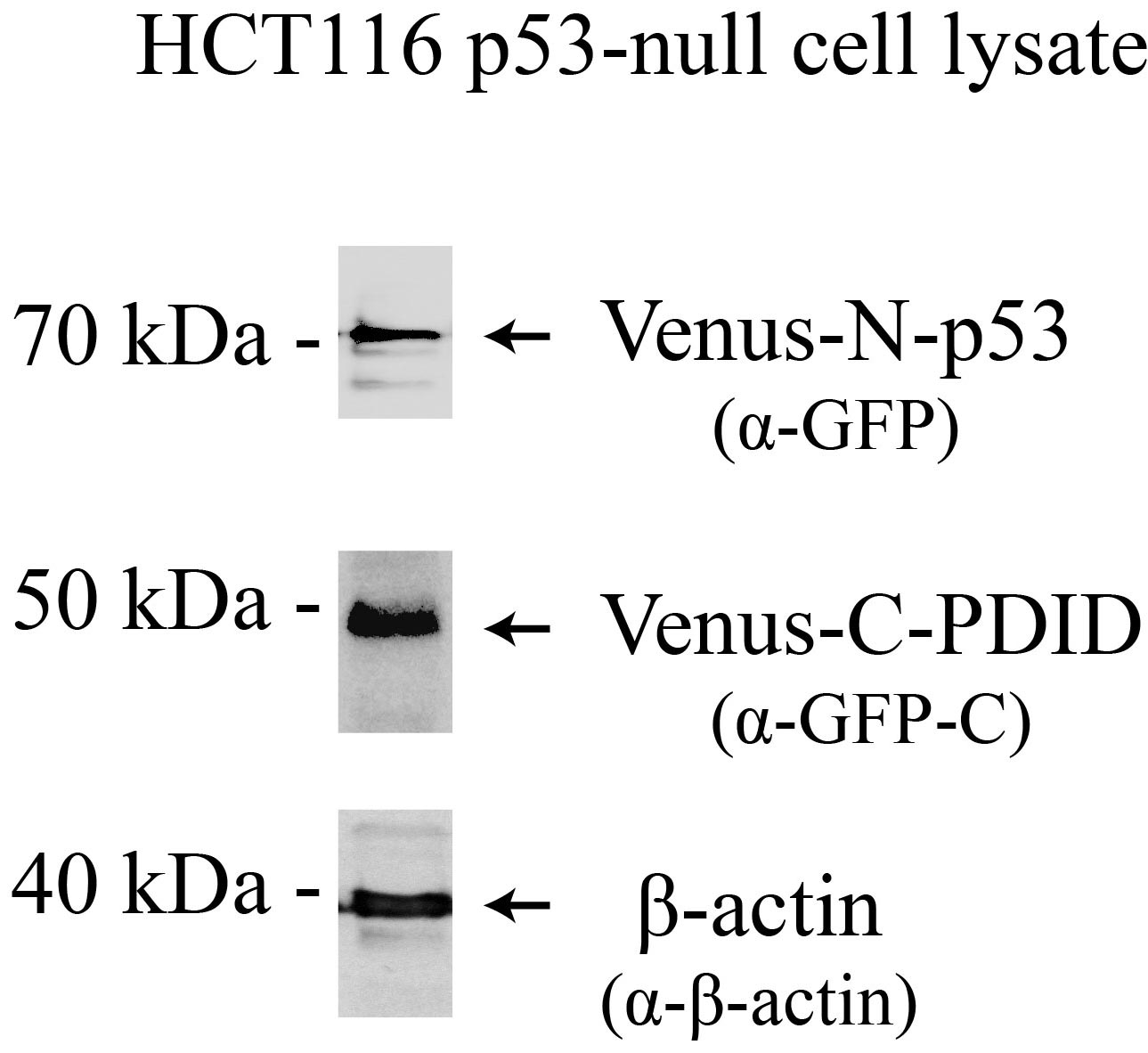
Figure 2. Venus-N-p53 and Venus-C-PDID protein expression. Western blot analysis of Venus protein expression in p53-null (p53-/-) HCT116 cells, 24 hours post-transfection. Antibodies: Venus-N-p53, Venus-C-PDID, and β-Actin.
- Wash cells for 5 min with 2 ml of 1x PBS on a rocker at a speed of 10-20 rpm, total three times.
Note: This step is important to reduce the background signals of Hoechst 33258 due to non-specific adherence of transfection DNA deposits to the plate surfaces. - Prepare for cell imaging:
For live-cell imaging:
Note: Perform steps a to c (see below) on a rocker at the speed of 10-20 rpm.
Note: Avoid center glass area when pipetting solutions at all steps.- Stain DNA in the nucleus with Hoechst 33258, 2 ml (5 μg/ml), for 30 min at room temperature.
Note: For multiple dishes, prepare one dish at a time before next Hoechst 33258 staining so there is enough time for fluorescence microscope visualization.
- Remove staining solution and wash cells for 5 min with 2 ml of 1x PBS, total three times.
- Add 1 ml of 1x PBS for fluorescence detection.
- Visualize fluorescence signals under a fluorescence microscope.
- Acquire lower magnification images and then higher magnification images of bright field, Venus, and Hoechst 33258.
Note: This step may take up to 30 to 60 min depending on adjusting position/focus and higher resolution of images desired.
- Typical results are shown in Figure 3.
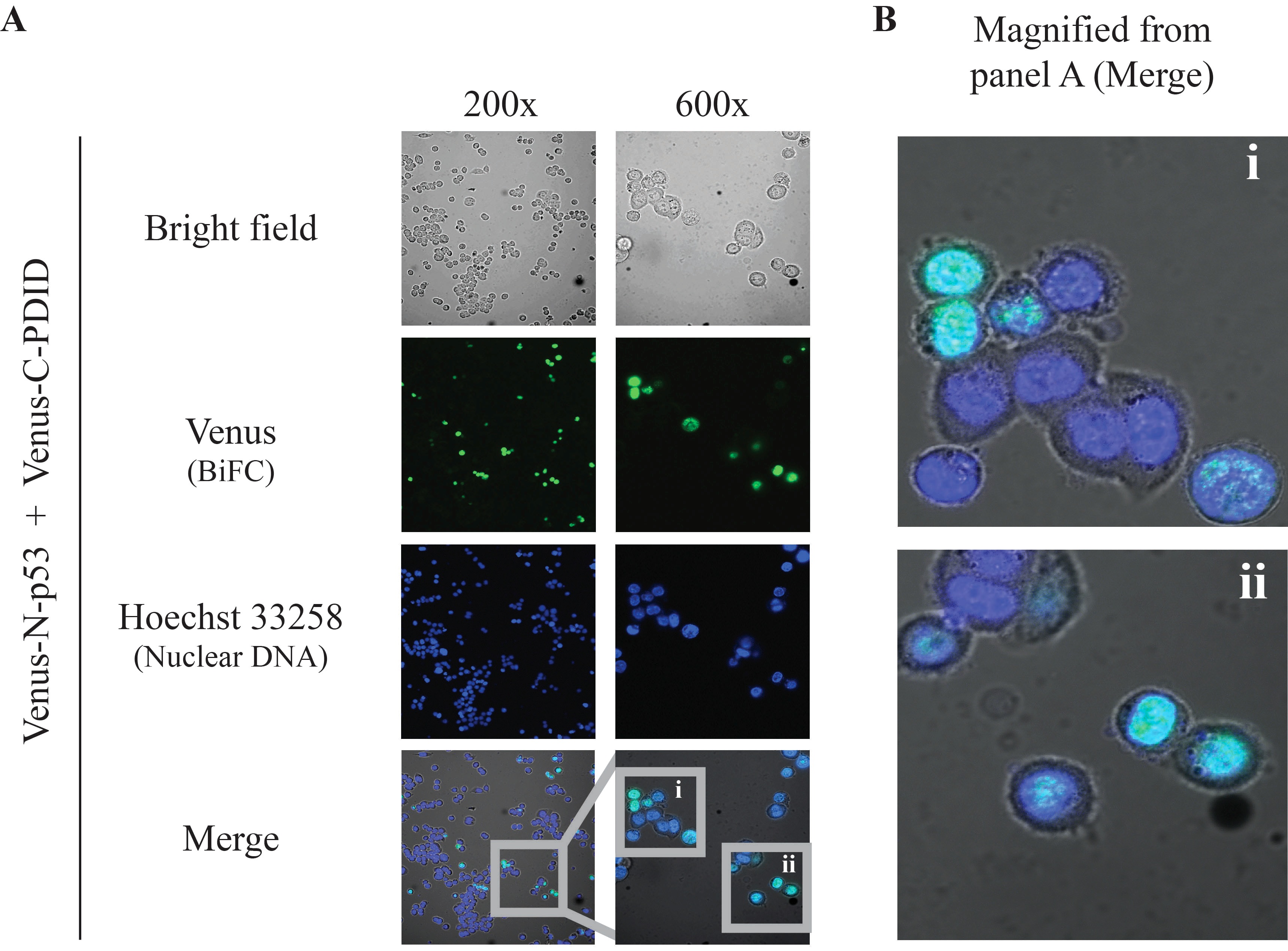
Figure 3. BiFC results. Direct visualization of p53-PDID interaction in vivo by BiFC live-cell imaging performed with p53-null (p53-/-) HCT116 cells transiently expressing Venus-N-p53 and Venus-C-PDID. A. Two different magnification images were obtained by using the 10x ocular lens in combination with a 20x or 60x objective lens. A 20x objective lens is typically used for observing a large number of cells and providing a general glimpse of cellular localization, whereas a 60x objective lens allows more detailed localization within subcellular compartments (Kerppola, 2006). Merge: combined Venus (pseudo-colored green), Hoechst 33258 (pseudo-colored blue), and bright field signals. B. Magnified images of p53-PDID interaction shown in A (i and ii) from 600x images. The BiFC signals co-localize with nuclear DNA staining (Hoechst 33258) as presented in pseudo-colored cyan. Images were obtained by Nikon Eclipse TE-2000E/C1 confocal fluorescence microscopy using NIS Element Basic Research software and further processed by Nikon EZ-C1 software.
Note: Fixed cell images are virtually the same as live-cell imaging (data not shown).
Note: Perform steps a to k (see below) on a rocker at a speed of 10-20 rpm.
Note: Avoid center glass area when pipetting solutions at all steps.- Fix cell with 1 ml 3.7% formaldehyde in PBS for 15 min at room temperature.
Note: 3.7% formaldehyde must be freshly prepared.
- Remove formaldehyde solution and wash fixed cells 5 min with 2 ml of 1x PBS, total three times.
- Remove wash solution and incubate fixed cells with 2 ml of 1x PBS containing 0.25% Triton X-100 to permeabilize cells for 30 min at room temperature.
- Wash cells for 5 min with 2 ml of 1x PBS, total three times.
Note: Skip antibody incubation procedures (steps e to i) if only signals from Venus and Hoechst 33258 (i.e., without fluorophore-conjugated antibody amplification) are needed. However these steps are helpful for verifying protein expression in transfected cells.
- Incubate permeabilized cells with 2 ml of PBSTB for 30 min to block non-specific antibody binding.
- Replace PBSTB with primary antibody (against protein of interest or GFP) in 1 ml of PBSTB for 1 h at room temperature or overnight at 4 °C.
Note: Start with 1:500 antibody dilution to determine the best condition. Two primary antibodies can be used at the same time.
- Wash cells for 5 min with 2 ml of 1x PBS, total three times.
- Incubate with secondary antibody (conjugated with fluorophores) in 1 ml of PBSTB for 1 hour at room temperature in the dark (wrap with aluminum foil).
Note: Start with 1:1,000 antibody dilution to determine best condition.
- Wash cells for 5 min with 2 ml of 1x PBS, total three times.
- Stain cell nucleus DNA with Hoechst 33258, 2 ml (0.5 μg/ml), for 10 min.
- Wash cells for 5 min with 2 ml of 1x PBS, total three times.
- Add 1 ml of 1x PBS for fluorescence detection.
- Visualize reconstituted fluorescence signal under a fluorescence microscope.
- Acquire lower magnification images and then higher magnification images of bright field, Venus, Hoechst 33258, or other signals from fluorophore-conjugated secondary antibody.
- Stain DNA in the nucleus with Hoechst 33258, 2 ml (5 μg/ml), for 30 min at room temperature.
- When needed, permanent preservation of samples can be done by the following steps:
- Remove PBS and use razor blade to separate the bottom glass (i.e., coverslip) from the petri dish (see Figure 4A).
Note: Do not touch the cell-attached side in the center circle of coverslip.
- Drop 50 μl of mounting medium on a microscope slide (see Figure 4B).
- Gently tilt the coverslip (see Figure 4C), with the cell-attached side facing down, to mount with mounting medium on a microscope slide (see Figure 4D).
- Gently press coverslip in the center with a pipet tip to remove air bubbles (see Figure 4E) and remove excess mounting medium around the edges with a paper towel (see Figure 4F).
- Mark microscope slides and seal coverslip with nail polish around the edges for 15 min, or until dry, to prevent sample movement and drying (see Figure 4G).
- Store at -20 °C or 4 °C in the dark.
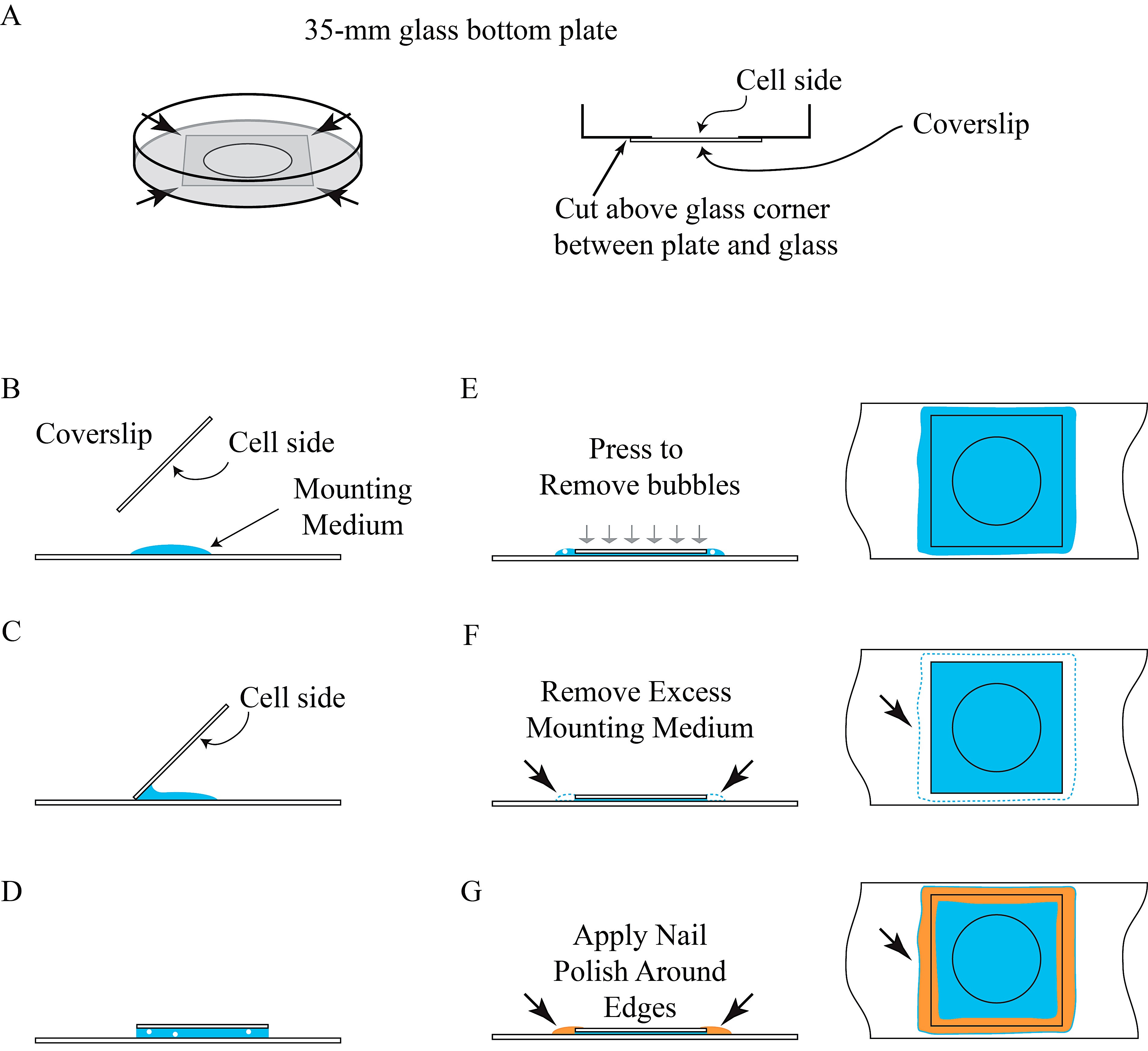
Figure 4. Illustration of permanent preservation of BiFC samples
- Remove PBS and use razor blade to separate the bottom glass (i.e., coverslip) from the petri dish (see Figure 4A).
Recipes
- 10x PBS (1 L)
80 g NaCl
2 g KCl
21.7 g Na2HPO4.7H2O
2 g KH2PO4
Add ddH2O to 1 L
Autoclave and stored at room temperature
Dilute with ddH2O to make 1x PBS (autoclave required, then store at room temperature)
- 3.7% Formaldehyde (freshly prepared for 10 ml)
1 ml 37% Formaldehyde
1 ml 10x PBS
8 ml sterilized ddH2O
- PBSTB (freshly prepared for 100 ml)
10 ml 10x PBS
250 μl Triton X-100
1 g BSA
Add sterilized ddH2O to 100 ml
- Hoechst 33258
Dissolve in 1x PBS to make final stock concentration 10 mg/ml
Dilute to desired concentration with 1x PBS
Stored at 4 °C
Acknowledgments
We thank Dr. Shwu-Yuan Wu for technical help and discussions during the development and writing of this protocol. The protocol detailed here was extended primarily from the procedures described in Wu et al. (2013). This work was supported in part by NIH grants (CA103867 and CA124760), CPRIT grants (RP110471 and RP120340), and a Welch Foundation grant (I-1805).
References
-
- Ding, Z., Liang, J., Lu, Y., Yu, Q., Songyang, Z., Lin, S. Y. and Mills, G. B. (2006). A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc Natl Acad Sci U S A 103(41): 15014-15019.
- Gully, C. P., Velazquez-Torres, G., Shin, J. H., Fuentes-Mattei, E., Wang, E., Carlock, C., Chen, J., Rothenberg, D., Adams, H. P., Choi, H. H., Guma, S., Phan, L., Chou, P. C., Su, C. H., Zhang, F., Chen, J. S., Yang, T. Y., Yeung, S. C. and Lee, M. H. (2012). Aurora B kinase phosphorylates and instigates degradation of p53. Proc Natl Acad Sci U S A 109(24): E1513-1522.
- Hu, C. D., Chinenov, Y. and Kerppola, T. K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9(4): 789-798.
- Kerppola, T.K. (2006). Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protocols 1(3): 1278-1286.
- Kerppola, T. K. (2013). Design of fusion proteins for bimolecular fluorescence complementation (BiFC). Cold Spring Harb Protoc 2013(8): 714-718.
- Morell, M., Espargaro, A., Aviles, F.X., and Ventura, S. (2008). Study and selection of in vivo protein interactions by coupling bimolecular fluorescence complementation and flow cytometry. Nat Protocols 3(1): 22-33.
- Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K. and Miyawaki, A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20(1): 87-90.
- Remy, I. and Michnick, S.W. (2007). Application of protein-fragment complementation assays in cell biology. BioTechniques 42(2): 137-145.
- Wu, S. Y., Lee, A. Y., Lai, H. T., Zhang, H. and Chiang, C. M. (2013). Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell 49(5): 843-857.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lai, H. and Chiang, C. M. (2013). Bimolecular Fluorescence Complementation (BiFC) Assay for Direct Visualization of Protein-Protein Interaction in vivo. Bio-protocol 3(20): e935. DOI: 10.21769/BioProtoc.935.
Category
Cell Biology > Cell imaging > Fluorescence
Molecular Biology > Protein > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link