- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Drug Sensitivity Assay of Xanthomonas citri subsp. citri Using REMA Plate Method
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.861 Views: 10924
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Survival of Wildtype and Mutant Escherichia coli in Soil
Yinka Somorin and Conor O'Byrne
Jul 20, 2017 8832 Views

Plaque Assay to Determine Invasion and Intercellular Dissemination of Shigella flexneri in TC7 Human Intestinal Epithelial Cells
Atin Sharma and Andrea Puhar
Jul 5, 2019 6209 Views

Shipment of Cyanobacteria by Agarose Gel Embedding (SCAGE)—A Novel Method for Simple and Robust Delivery of Cyanobacteria
Phillipp Fink [...] Karl Forchhammer
Dec 5, 2024 1393 Views
Abstract
Resazurin Microtiter Assay (REMA) is a simple, rapid, reliable, sensitive, safe and cost-effective measurement of cell viability. Resazurin detects cell viability by converting from a nonfluorescent dye to the highly red fluorescent dye resorufin in response to chemical reduction of growth medium resulting from cell growth (Palomino et al., 2002). The REMA assay can be used as a fluorogenic oxidation-reduction indicator in a variety of cells, including bacteria, yeast and eukaryotes (Silva et al., 2013).
Materials and Reagents
- Chemicals: Synthetic esters of gallic acids (Ximenes et al., 2010)
- Bacterial strain: Wild type Xanthomonas citri subsp citri strain 306 (Schaad et al., 2005)
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- Kanamycin (Sigma-Aldrich, catalog number: K4000 )
- Luria-Bertani broth (LB) culture medium
- Resazurin sodium salt (Sigma-Aldrich, catalog number: R7017 )
Equipment
- 96-well plate, polystyrene, with clear flat bottom wells (Greiner Bio-one, catalog number: 655101 )
- SPECTRAfluor Plus (Tecan) microfluorimeter
- Multichannel pipetman (Eppendorf)
Procedure
- Prepare stock solutions of chemicals (dried-powder samples) dissolving in 10% in DMSO (diluted in sterile water).
- Add 100 μl of water to columns 1 and 12 to avoid evaporation (Table 1).
- Dilute the stock solutions in LB medium directly in a 96-well plates using a 2-fold scheme (final volume of 100 μl per a well); after serial dilution, the most concentrated sample should have maximum 1% DMSO.
- Cells were grown in LB medium at 30 °C under rotation (200 rpm) until OD600 0.6 ( log phase).
- Add 10 μl of bacterial inoculum (standardized to 105 CFU/well).
- Negative control: 1% DMSO dissolved in LB.
- Positive control: Kanamycin at 15.6 μg/ml.
Table 1. Example for setup of REMA 96-well assay plate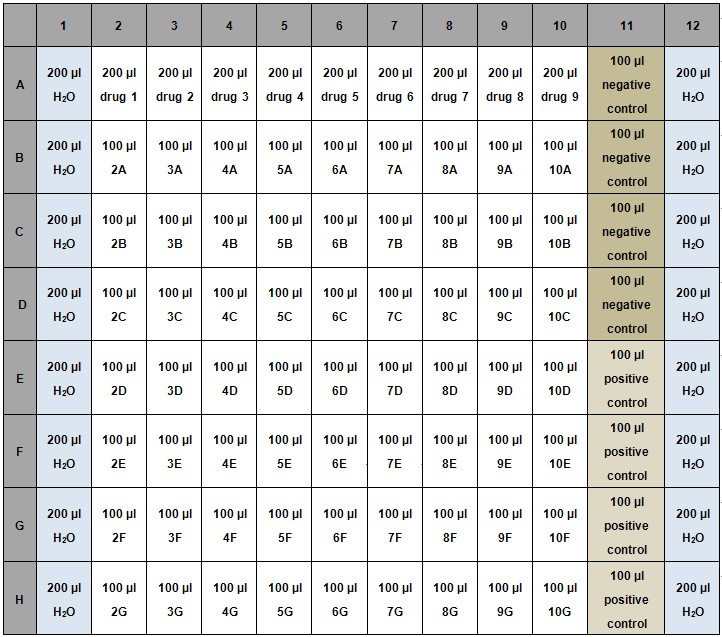
- Negative control: 1% DMSO dissolved in LB.
- Incubate the test plates at 30 °C for 6 h.
- Add 15 μl of a 0.01% (w/v) resazurin solution, and incubate at 30 °C for 2 h.
- Measure fluorescence at 530 nm (excitation) and 590 nm (emission) using a fluorescence scanning.
- Percentage of inhibition is defined as:
[(average FU negative control) - (average FU test)]/(average FU negative control) x 100
FU: Fluorescence Units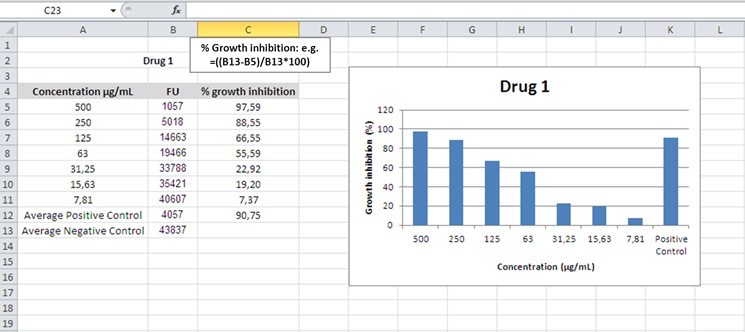
Figure 1. Example for calculation of growth inhibition
Note: Three independent experiments should be conducted, and the data is used to construct plots of chemical concentration versus cell growth inhibition in order to determine the MIC* (Figure 1).
*The minimum inhibitory concentration (MIC) is defined as the lowest concentration of the antibiotic able to inhibit the growth of 90% of organisms.
Acknowledgments
This work was supported by FAPESP research grants 2004/09173-6, 2010/05099-7, and 2011/07458-7. This protocol was adapted from a previous work by Palomino et al. (2002).
References
- Palomino, J. C., Martin, A., Camacho, M., Guerra, H., Swings, J. and Portaels, F. (2002). Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46(8): 2720-2722.
- Schaad, N. W., Postnikova, E., Lacy, G. H., Sechler, A., Agarkova, I., Stromberg, P. E., Stromberg, V. K. and Vidaver, A. K. (2005). Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and "var. fuscans" of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst Appl Microbiol 28(6): 494-518.
- Silva, I. C., Regasini, L. O., Petronio, M. S., Silva, D. H., Bolzani, V. S., Belasque, J., Jr., Sacramento, L. V. and Ferreira, H. (2013). Antibacterial activity of alkyl gallates against Xanthomonas citri subsp. citri. J Bacteriol 195(1): 85-94.
- Ximenes, V. F., Lopes, M. G., Petronio, M. S., Regasini, L. O., Silva, D. H. and da Fonseca, L. M. (2010). Inhibitory effect of gallic acid and its esters on 2,2'-azobis(2-amidinopropane)hydrochloride (AAPH)-induced hemolysis and depletion of intracellular glutathione in erythrocytes. J Agric Food Chem 58(9): 5355-5362.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Silva, I. C. and Ferreira, H. (2013). Drug Sensitivity Assay of Xanthomonas citri subsp. citri Using REMA Plate Method. Bio-protocol 3(16): e861. DOI: 10.21769/BioProtoc.861.
Category
Microbiology > Microbial cell biology > Cell viability
Cell Biology > Cell viability > Cell death
Cell Biology > Cell staining > Whole cell
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link