- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Northwestern Blot of Protein-RNA Interaction from Young Rice Panicles
Published: Vol 3, Iss 7, Apr 5, 2013 DOI: 10.21769/BioProtoc.625 Views: 16251
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Immunoprecipitation Using Nicotiana benthamiana Transient Expression System
Fang Xu [...] Xin Li
Jul 5, 2015 23173 Views

Mating Based Split-ubiquitin Assay for Detection of Protein Interactions
Wijitra Horaruang and Ben Zhang
May 5, 2017 17304 Views

Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 1996 Views
Abstract
The northwestern assay is employed to study the interaction between protein and RNA. The RNA binding proteins tend to bind to different kinds of RNA through either known domains or unknown sequences of proteins. Rice LGD1 recombinant protein, a grass-specific novel protein with RNA binding sequences in its C-terminal, was used to probe its function as an RNA binding protein. The LGD1 comprising von Willebrand factor type-A domain (vWA), coiled-coil and nuclear localization signal (NLS) is a class of protein that localizes both in the nucleus and cytoplasm. Although LGD1 does not contain any putative RNA binding domains, we could find high-affinity RNA binding residues at the C-terminus using ‘RNABindR’ prediction software (Terribilini et al., 2007). The LGD1 recombinant protein, purified from bacteria, somehow forms both dimer and monomer even under denaturing conditions. However, only the dimeric form is able to bind to total and mRNAs. Due to its reproducibility and reliability, we believe that this protocol can be used across different organisms.
Keywords: LGD1Materials and Reagents
- Young panicle tissues of wild type (Oryza sativa L. ssp. Japonica cultivar Tainung67) plants
- Tissues were collected from young panicles while they were in secondary branch initiation
- Recombinant GST-LGD1 protein from E. coli (BL21)
- pGEX-4T-1 GST expression vector (GE Healthcare, catalog number: 28-9545-49 )
- Total RNA isolation kit - Trizol (Life Technologies, InvitrogenTM, catalog number: 15596-026 )
- mRNA purification kit - Oligotex mRNA Mini Kit (QIAGEN, catalog number: 72022 )
- RNA 5'-end Labeling kit (T4 Polynucleotide Kinase) (Thermo Fisher Scientific, catalog number: EK0031 )
- FastAP Thermosensitive Alkaline Phosphatase (Thermo Fisher Scientific, catalog number: EF0651 )
- [γ-32P]-ATP–for isotope labeling
- PVDF Immobilon-P membrane (EMD Millipore, catalog number: IPVH15150 )
- G-50 gel filtration dye terminator removal column (Geneaid, Taiwan, catalog number: CG050 )
- Primary anti-GST antibody (Bioman, Taiwan, catalog number: GST001M ); HRP-conjugated secondary antibody (Thermo Fisher Scientific, catalog number: SA1-9510 )
- SuperSignal West Pico Chemiluminescence detection system (Thermo Fisher Scientific, catalog number: 34087 )
- Blocking buffer (see Recipes)
Equipment
- Gel electrophoresis (Bio-Rad Laboratories, catalog number: 165-8005EDU )
- SNAP i.d.® 2.0 protein detection system
- Equipment for phosphor imaging (Bio-Rad Laboratories, catalog number: 170-9460 )
Procedure
- RNA isolation and end-labeling of total and mRNA
- Total RNA was isolated from young panicles of wild type plants with Trizol kit.
- ~0.5 g tissue was used to isolate ~500 μg of total RNA and 200 μg total RNA was used to purify ~2 μg mRNA using Oligotex mRNA Mini Kit. Nanodrop was used for final RNA quantitation (~2 μg/μl for total RNA; ~20 ng/μl for mRNA).
- 5 μg total RNA and 1 μg mRNA were first treated with FastAP Thermosensitive Alkaline Phosphatase (10 U) at 37 °C for 1 h to remove 5’-phosphate group according to manufacturer’s protocol.
- The total and mRNAs were end-labeled with [γ-32P]-ATP using 1 μl (10 U) T4 Polynucleotide kinase at 37 °C for 30 min.
- Labeled RNAs were separated from unincorporated label by gel filtration on Sephadex G-50 column.
- Total RNA was isolated from young panicles of wild type plants with Trizol kit.
- Northwestern blot assay
- GST-LGD1 fusion protein was extracted from E. coli harboring pGEX-4T-1 GST expression vector according to manufacturer’s protocol.
- The proteins were separated on 12% SDS PAGE under denaturing conditions, and transferred to PVDF Immobilon-P membrane and detected using the SNAP i.d. protein detection system as follows.
- For the Western blot, the primary anti-GST antibody (1:2,000) and secondary HRP (horseradish peroxidase)-conjugated antibody (1:3,000) were used for the enhanced chemiluminescence (ECL) detection.
- SuperSignal West Pico Chemiluminescence was employed to detect western signals according to the manufacturer’s instructions.
- For the Northwestern, recombinant proteins separated on 12% SDS PAGE under denaturing conditions were transferred to PVDF Immobilon-P membrane.
- The proteins were then renatured on the blots overnight at 4 °C in a renaturation buffer containing 0.1 M Tris HCl (pH 7.5) and 0.1% (v/v) NP-40.
- Blots were washed 4 times 15 min each in renaturation buffer and incubated 5 min at room temperature in blocking buffer.
- Blots were hybridized overnight at 4 °C in blocking buffer (without BSA) in the presence of labeled total and mRNAs.
- Finally, the blots were washed 4 times 5 min each in blocking buffer (without BSA and Triton) and autoradiographed to obtain the signal as seen in Figure 1.
- GST-LGD1 fusion protein was extracted from E. coli harboring pGEX-4T-1 GST expression vector according to manufacturer’s protocol.
Representative data
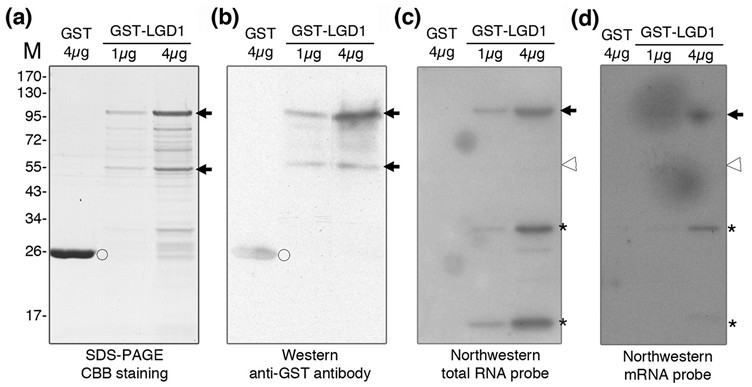
Figure 1. Northwestern blot analysis of LGD1 recombinant protein. The recombinant GST-LGD1 was purified from E. coli and analyzed through SDS-PAGE, western and northwestern blots. (a) Coomassie Brilliant Blue (CBB) staining shows two major bands (arrows). The size of the lower band is ~56 kD (monomer) and upper band is ~112 kD (homodimer). (b) Anti-GST antibody recognizes the GST-LGD1 fusion protein (arrows) as well as GST (open circle). Northwestern blot shows the total RNA. (c) and mRNA (d) isolated from young panicle tissues bind to upper band (arrows), but not to the lower band (open arrowhead). Asterisks indicate RNA binding to degraded form of LGD1.
Recipes
- Blocking buffer
10 mM Tris HCl (pH 7.5)
5 mM Mg acetate
2 mM DTT
5 % (w/v) BSA
0.01 % (v/v) Triton X-100
Acknowledgments
We greatly appreciate the contributions of Drs Yue-Ie Hsing, Chyr-Guan Chern, Ming-Jen Fan and Su-May Yu to generate the TRIM database. We also thank Drs Ko Shimamoto (Nara Institute of Science and Technology, Japan), for sharing the pANDA-RNAi vector, and Su-May Yu (Institute of Molecular Biology, Academia Sinica), for excellent support with rice transgenic experiments. We also thank Ms Lin-Yun Kuang (Transgenic Plant Laboratory, Academia Sinica) for assistance in particle bombardment. We are grateful to Ms. Krisa Fredrickson for her English editing. This work was supported by research grants from Academia Sinica (Taiwan), the National Science and Technology Program for Agricultural Biotechnology (NSTP/AB, 098S0030055-AA, Taiwan), the National Science Council (98-2313-B-001-001-MY3, Taiwan) and the Li Foundation (USA) to Guang-Yuh, Jauh.
References
- Uyttewaal, M., Mireau, H., Rurek, M., Hammani, K., Arnal, N., Quadrado, M. and Giege, P. (2008). PPR336 is associated with polysomes in plant mitochondria. J Mol Biol 375(3): 626-636.
- Terribilini, M., Sander, J. D., Lee, J. H., Zaback, P., Jernigan, R. L., Honavar, V. and Dobbs, D. (2007). RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res 35(Web Server issue): W578-584.
- Thangasamy, S., Chen, P. W., Lai, M. H., Chen, J. and Jauh, G. Y. (2012). Rice LGD1 containing RNA binding activity affects growth and development through alternative promoters. Plant J 71(2): 288-302.
- Uyttewaal, M., Mireau, H., Rurek, M., Hammani, K., Arnal, N., Quadrado, M. and Giege, P. (2008). PPR336 is associated with polysomes in plant mitochondria. J Mol Biol 375(3): 626-636.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Thangasamy, S. and Jauh, G. (2013). Northwestern Blot of Protein-RNA Interaction from Young Rice Panicles. Bio-protocol 3(7): e625. DOI: 10.21769/BioProtoc.625.
Category
Plant Science > Plant biochemistry > Protein > Interaction
Biochemistry > Protein > Interaction > Northwestern blot
Biochemistry > RNA > RNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










