- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Using BODIPY FL-Sphingolipid Analogs to Study Sphingolipid Metabolism in Mouse Embryonic Stem Cells
Published: Vol 12, Iss 22, Nov 20, 2022 DOI: 10.21769/BioProtoc.4555 Views: 2592
Reviewed by: Julie WeidnerAgnieszka ZienkiewiczYu Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2901 Views

Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient
Vikram Aditya [...] Wei Yue
Apr 5, 2025 2857 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views
Abstract
Sphingolipids are important structural components of cellular membranes. They also function as prominent signaling molecules to control a variety of cellular events, such as cell growth, differentiation, and apoptosis. Impaired sphingolipid metabolism, particularly defects in sphingolipid degradation, has been associated with many human diseases. Fluorescence sphingolipid analogs have been widely used as efficient probes to study sphingolipid metabolism and intracellular trafficking in living mammalian cells. Compared with nitrobenzoxadiazole fluorophores (NBD FL), the boron dipyrromethene difluoride fluorophores (BODIPY FL) have much higher absorptivity and fluorescence quantum. These features allow more intensive labeling of cells for fluorescence microscopy imaging and flow cytometry analysis. Here, we describe a protocol employing BODIPY FL-labeled sphingolipid analogs to elucidate sphingolipid internalization, trafficking, and endocytosis in mouse embryonic stem cells.
Graphical abstract:
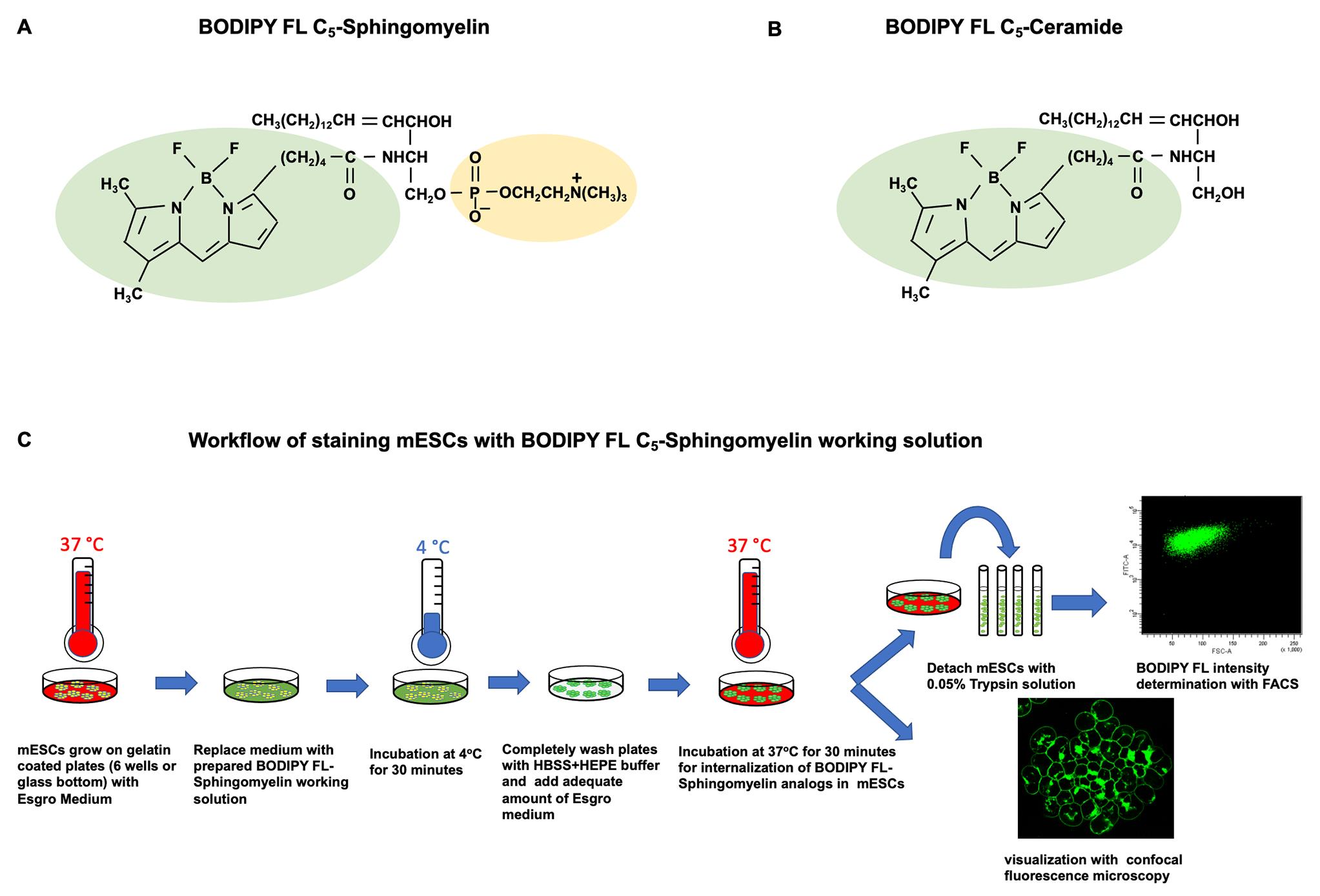
Background
Sphingolipids are a group of structurally diverse lipids, first discovered from brain extract in the 1880s. Globally, they act as essential components of the plasma membrane in almost all types of vertebrate cells (Schnaar and Kinoshita, 2015). All sphingolipids share a common sphingoid-based backbone (Figure 1A), which acts as the structural foundation for all sphingolipid derivatives. Adding one fatty acid to this sphingosine base forms ceramide, the central molecule in sphingolipid biology (Figure 1B). Further adding a phosphoryl choline or phosphoryl ethanolamine group to ceramide makes sphingomyelin (SM), the most abundant mammalian sphingolipid (Figure 1C and 1D) (Fan et al., 2021).

Figure 1. Chemical structure of sphingolipids. General chemical structure of (A) sphingosine backbone, (B) ceramide, which has an additional fatty acid molecule (highlighted in red) attached to the sphingosine backbone, and (C-D) the two most common types of sphingomyelin, which have one (C) phosphocholine or (D) phosphoethanolamine group (highlighted in blue) attached to the ceramide.
In mammalian cells, the endoplasmic reticulum (ER), Golgi complex, and plasma membrane are key subcellular locations hosting sphingolipid metabolism. They accommodate the majority of the two most important and abundant types of sphingolipids: ceramide and SM. Ceramide is the central node of the sphingolipid metabolism network. There are two endogenous ceramide synthesis pathways: the first one is the de novo synthesis in ER. The newly synthesized ceramide is then transported into the Golgi complex for further conversion to more complex forms of sphingolipids, such as SM; the second one is the regeneration from complex sphingolipids in the Golgi complex and plasma membrane, by a class of specific hydrolases and phosphodiesterases, including the plasma membrane-bound sphingomyelin phosphodiesterases (Gault et al., 2010; Heinz et al., 2015; Fan et al., 2021).
The sphingolipids are not only important in supporting physical microdomain structures of mammalian cell membranes, but also act as prominent signaling molecules controlling a number of cellular events, such as cell growth, differentiation, and apoptosis (Laude and Prior, 2004). The significance of sphingolipids for human health is best demonstrated by human neural degenerative diseases, including Alzheimer’s, Parkinson’s, and Niemann-Pick disease. These neural degenerative diseases are known to be associated with dysregulation or disturbance of key sphingolipids, such as SM and ceramide (Bienias et al., 2016; Alessenko and Albi, 2020). The elucidation of sphingolipid internalization, trafficking, and endocytosis in live mammalian cells, therefore, is key to the understanding of the underlying mechanisms of these human diseases.
Lipid analogs generated by replacing naturally occurring fatty acids in complex lipids with artificial short-chain fatty acids containing fluorophores (FL) have been widely used to study membrane lipid trafficking in mammalian cells (Koval and Pagano, 1991; Hoekstra and Kok, 1992; Rosenwald and Pagano, 1993). For example, the fluorophore boron dipyrromethene difluoride (BODIPY)- (Figure 2A) or nitrobenzoxadiazole (NBD)-labeled SMs and ceramides (Figure 2B and 2C), have been widely applied as molecular probes for lipid trafficking in live mammalian cells. These artificial lipid analogs equipped with BODIPY or NBD fluorophores can be readily integrated into cellular membranes through spontaneous lipid transfer from exogenous sources. Further intracellular distribution of those lipid analogs, or metabolites derived from them, can be visualized and quantified with high resolution fluorescence microscopy (confocal laser scanning microscopy [CLSM]) or fluorescence-activated cell sorting flow cytometry (FACS) (Pagano and Chen, 1998).
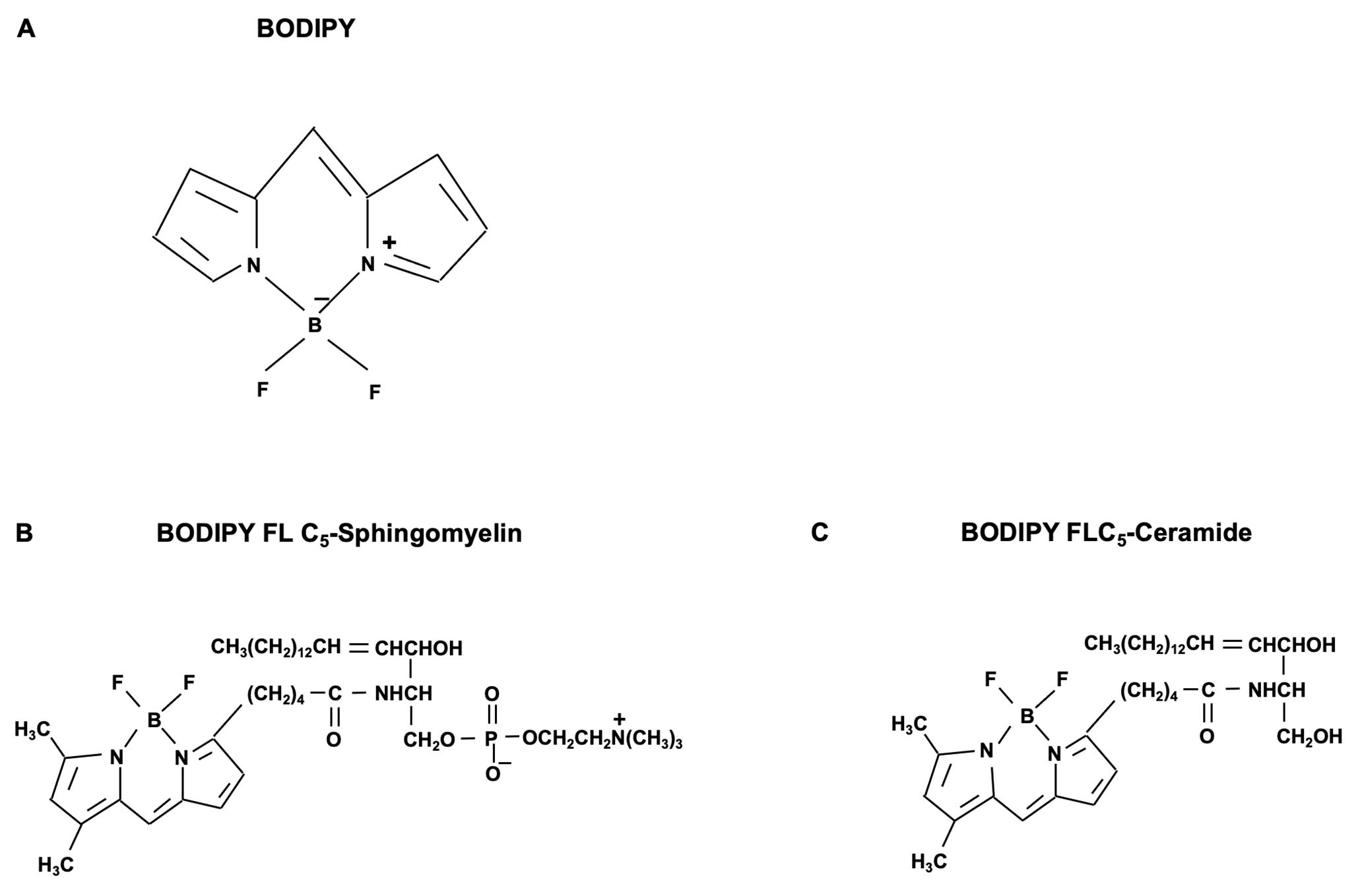
Figure 2. Chemical structure of sphingolipid analogs with BODIPY fluorophore. The chemical structure of (A) boron dipyrromethene difluoride chelate (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene) fluorophore (BODIPY FL), (B) BODIPY FL C5-Sphingomyelin, and (C) BODIPY FL C5-Ceramide.
Both BODIPY and NBD are widely used fluorophores. Compared with NBD, the BODIPY fluorophore has several advantages for studying lipid metabolism and trafficking, specifically in live cells. First, BODIPY has stronger fluorescence and is more photostable than NBD (Johnson et al., 1991). Second, BODIPY’s structure is less polar than that of NBD; therefore, it is more efficiently anchored in the bilayer of the mammalian cell membrane. In contrast, NBD easily loops back to the water/bilayer interface, which may interfere with integration of lipid analogs into the plasma membrane (Chattopadhyay and London, 1987; Wolf et al., 1992; Pagano and Chen, 1998). Third, the emission wavelength of BODIPY fluorescence ranges from 450 (green) to 650 (red) with increased concentration in cells (Pagano et al., 1991). This special feature makes the distribution of a particular BODIPY-labeled lipid analog and its metabolites dynamically visible, allowing easy observation and quantification in live cells under fluorescence microscopy (Pagano et al., 1991).
We recently investigated the importance of sphingolipid metabolism in neural differentiation of mouse embryonic stem cells (mESCs) using BODIPY FL sphingomyelin (Fan et al., 2021). Therefore, this protocol describes the application of commercially available BODIPY FL sphingomyelin analogs to visualize and quantify internalization, trafficking, and endocytosis of sphingolipids in mESCs, by FACS and CLSM. The protocol includes detailed methods for preparation of BODIPY FL sphingomyelin analog probes and staining of mESCs using these analogs, for FACS and CLSM analyses. We expect that this protocol will have a wide range of applications in sphingolipid research.
Things to consider before starting
Choice of BODIPY FL sphingolipids
There are many commercially available BODIPY FL sphingolipid analogs, containing different lengths of fatty acid chains or different substitutions on the BODIPY moiety to modify its spectral properties. A five-carbon fatty acid, 5-(5,7-dimethyl-BODIPY)-1-pentanoic acid, has been previously reported to be the most effective one for most applications (Pagano et al., 2000). Moreover, the spectral properties of C5-BODIPY lipid analogs are not sensitive to either pH or membrane potential and curvature (Karolin et al., 1994; Chen et al., 1997).
The excitation and emission of bright green-fluorescein generated by the commercially available BODIPY FL dye (Invitrogen) are similar to those of fluorescein (FITC) or Invitrogen Alexa Fluor 488 dye. However, compared with fluorophores Alexa Fluor 488 dye and FITC, BODIPY FL dye has higher extinction coefficient and increased fluorescence quantum yield, and is insensitive to solvent polarity and pH change. Also, due to its hydrophobic properties and long excited-state lifetime (typically, 5 nanoseconds or longer), BODIPY FL dye is very effective in labeling cellular lipids, specifically membrane lipids, and useful for fluorescence polarization-based assays (referred in the commercial product manual of Invitrogen D3522).
Expected subcellular locations of BODIPY FL sphingolipid analogs
The distribution of N-[5-(5,70-dimethyl BODIPY)-1-pentanoyl]-D-erythro-sphingosine (C5-DMB-Cer), a BODIPY FL ceramide analog, and its metabolites during endocytosis in human skin fibroblast cells has been previously traced (Pagano et al., 1991; Pagano and Chen, 1998). In those cells, this fluorescent lipid analog starts to be integrated into the plasma membrane and is then exclusively present in the outer leaflet of the plasma membrane bilayer at low temperature (commonly, at 4 °C) (Martin and Pagano, 1987; 1994; Chen et al., 1997). During subsequent temperature rising to 37 °C, this lipid analog starts to be internalized into lipid vesicles throughout the cytoplasm (Koval and Pagano, 1989; 1990). The whole internalization process is highly sensitive to low temperature, due to its strict energy dependence.
As soon as the internalization is activated by high temperature incubation, the internalized fluorescent lipid analogs can either return intact (recycle) back to the plasma membrane, or be transported to lysosome storage, and further hydrolyzed by lysosomal sphingolipid hydrolases (Koval and Pagano, 1990; Mayor et al., 1993; Grassi et al., 2019). In this process, in addition to BODIPY FL in the endosomes, BODIPY FL-ceramide resulting from the hydrolysis of BODIPY FL-C5-DMB-Cer at the plasma membrane are also subsequently transported to the Golgi apparatus, because of its high affinity for ceramides (Pagano, 1990; Rosenwald and Pagano, 1993). Therefore, in mESCs, after the stable state of internalization has been reached upon completion of 30 min incubation at 37 °C, multiple cellular membrane structures—including plasma membrane, Golgi complex, ER, and endosomes—are expected to “lighten up” due to BODIPY fluorescence.
Cell type-specific distribution of BODIPY FL sphingolipid analogs
It is worth noting that different cell types cultured in different media have distinct patterns of sphingolipid metabolism. For instance, ESCs require some specific features to maintain their pluripotency and unique functions (Tanosaki et al., 2021). Therefore, the endocytosis and metabolism of sphingolipids, which can be reflected by the shape, color, and distribution of these lipid analogs and their metabolites in cells, may differ greatly in different cells.
Materials and Reagents
mESC culture
Sterile serological pipettes (Serological pipettes of 5 mL, 10 mL, 25 mL and 50 mL; Sarstedt, catalog numbers: 86.1253.001, 86.1254.001, 86.1685.001)
Sterile Corning® centrifuge tubes (50 mL and 15 mL; Millipore, catalog numbers: CLS430290, CLS430055)
Falcon® 5 mL Round Bottom Polystyrene Test Tube (with snap Cap for FACS analysis, Sterile; Falcon, catalog number: 352058)
FisherbrandTM Class B Amber Glass Threaded Vials (1.8 mL, 3.7 mL, 7.4 mL and16 mL; Fisher Scientific, catalog number: 03-339-23)
6 wells sterile cell culture dishes (NuncTM Cell-Culture Treated Multidishes; Thermo Fisher Scientific, catalog number:140675)
NuncTM Glass Bottom Dishes (Perform high quality imaging in the ease of a 35 mm dish; Thermo Fisher Scientific, catalog number:150682)
R1 mESC or ES-E14TG2a (E14) mESC lines (ATCC, catalog number: SCRC-1011, RRID: CVCL_2167; CRL-1821, RRID: CVCL_9108)
Gelatin powder (Gelatin from porcine skin; Millipore Sigma, catalog number: G1890)
ESGRO Complete Plus Grade medium: a commercially developed serum free complete basal medium. It contains a selective GSK3β inhibitor to enhance viability of mESCs and increase maintenance of the pluripotency (Millipore, catalog number: SF001)
Cytiva HyClone Dulbecco's Phosphate Buffered Saline liquid (DPBS buffer; Fisher Scientific, catalog number: SH3002802)
Water for cell culture (sterile-filtered, BioReagent, suitable for cell culture; Millipore Sigma, catalog number: W3500)
GibcoTM Trypsin-EDTA (0.05%), phenol red (Sigma-Aldrich, catalog number: 25300054)
Trypan Blue Solution, 0.4% (Thermo Fisher Scientific, catalog number: 15250061)
0.1% Gelatin solution (see Recipes)
Preparation of BODIPY FL-sphingomyelin stock and working solutions
BODIPYTM FL C5-Sphingomyelin (Invitrogen, catalog number: D3522)
Defatted bovine serum albumin (BSA; Roche, catalog number: 03117057001)
Hanks’ Balanced Salt solution (HBSS buffer solution; HBSS, no calcium, no magnesium, no phenol red; Gibco, catalog number: 14175095)
HEPES solution (1 M N-2-Hydroxyethylpiperazine-N′-2-ethane sulfonic acid in H2O, Gibco, catalog number: 15630106)
Chloroform (anhydrous, ≥99%, contains 0.5–1.0% ethanol as stabilizer; Millipore Sigma, catalog number: 288306)
Ethyl alcohol (Absolute alcohol, 200 proof anhydrous CAS#64-17-5; The Warner Graham Company)
70% ethanol (see Recipes)
Chloroform:ethanol (19:1 v/v) (see Recipes)
HBSS/HEPES buffer (pH7.4) (see Recipes)
Equipment
Cell counter (Countess 3 Automated Cell Counter; Thermo Fisher)
Analytical Nitrogen Evaporator (24 Position N-EVAP Nitrogen Evaporator; Organomation Associate, Inc; model: N-EVAPTM 112 #11250)
Forma series 3 water jacketed CO2 incubator (Thermo Fisher)
Avanti J-15R Centrifuge with GH-3.8/GH-3.8A rotor (Beckman Coulter)
Mini Vortex Mixer (Variable Speed) (Fisher Scientific)
Refrigerator (Whirlpool)
Zeiss LSM 780 UV inverted confocal microscope with AiryScan (Carl Zeiss Sports Optics, model: LSM 780)
Fluorescence-Activated Cell Sorting flow cytometer (BD Bioscience, Brand, model: BD LSRFortessa with HTS option)
Software
Zeiss Zen (Carl Zeiss Sports Optics, web address: https://www.binran.ru/files/ckp/fluorestsentnaya-mikroskopiya/CKP_LSM780_ZEN2010_Manual_ENG.pdf)
BDFACSDivaTM software version 8.0.1 (BD Bioscience, web address: https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software)
Procedures
Plate and culture mESCs
Dissolve gelatin powder in cell culture-grade water to the final concentration of 0.1% (w/v), then sterilize this solution by autoclave.
Apply an appropriate amount of 0.1% gelatin solution to cover the entire surface area of cell culture dishes, then incubate the dishes at room temperature (RT) for 30 min to coat the cell growth area.
After incubation, aspirate the gelatin solution and wash the coated cell culture dishes with PBS solution twice for 3 min each time, to completely remove the residual gelatin solution.
Note: All above procedures (2–3) must be performed in a sterile hood. The resulting gelatin-coated dishes will be set aside in PBS solution for fresh cell plating. They can also be dried then kept at RT for up to 1 month for future use (after rehydration with PBS solution).
Completely disassociate pre-cultured mESCs, by adding an appropriate amount of 0.05% trypsin (e.g., an approximate volume of 300 µL for each well of the 6-well plate) to cover the entire cell growth area, then incubate at 37 °C for 3 min. After cells are completely disassociated from dishes, wash them twice with 30 mL of PBS solution in 50-mL Corning tubes, to completely remove residual trypsin. Harvest cells by centrifugation at 300 × g for 3 min.
Note: mESC colonies must be completely disassociated with 0.05% trypsin (extra amount of trypsin and extended digestion time may be applied) to ensure all cells are evenly dispersed in cell suspension before re-plating. Failure to complete dissociate cells could induce differentiation of mESCs, and/or formation of big cell clumps that interfere with subsequent analyses.
Resuspend mESCs with the ESGRO growth medium, and plate cells on gelatin-coated 6-well cell culture plates for FACS analysis, or NuncTM Glass Bottom Dishes for CLSM visualization, at the density of approximately 5 × 104 cells/cm2. Incubate cells in a cell culture incubator at 37 °C with 95% humidity and 5% CO2. Cells will be stained with BODIPY FL-sphingomyelin analogs after their attachment to dishes, or after overnight incubation.
Note: mESCs are very sensitive to environmental perturbations, including chemical and physical stresses. Even very minor agitations may result in apoptosis, differentiation, or morphological changes of mESCs. Therefore, cell culture procedures must be performed in an extraordinarily careful and gentle manner.
Prepare the stock and working solutions of BODIPY FL C5-sphingomyelin analogs
Stock solution: Mix absolute chloroform and absolute ethanol at a ratio of 19:1 (v/v) (Recipe 3) inside a chemical safety cabinet. To prepare 1 mM BODIPY FL C5-sphingomyelin stock solution, directly inject an appropriate amount of the prepared chloroform/ethanol solvent into the original product vial containing the BODIPY FL C5-sphingomyelin powder. Shake vials until all powders are completely dissolved in the solvent, then transfer the 1 mM stock solution to amber glass round vials. This stock solution must be stored at -20 °C and strictly protected from light.
Working solution:
Dispense 50 µL of 1 mM BODIPY FL C5-sphingomyelin stock solution from step B1 into an amber glass round vial. Evaporate the organic solvent under a stream of nitrogen, by placing the nozzle (like a needle) of the nitrogen evaporator into the mouth of the amber glass round vial, and spraying the nitrogen gas toward the stock solution, until no liquid phase is observed. After the liquid phase has been completely dried off, add 200 µL of absolute ethanol into the amber glass round vial, to reconstitute the dried BODIPY FL C5-sphingomyelin powder.
To prepare 5 µM defatted BSA solution, dissolve an appropriate amount of defatted BSA into 10 mL of Hanks’ buffered salt solution containing 10 mM HEPES (HBSS/HEPES buffer, Recipe 4).
Add the prepared 200 μL of BODIPY FL C5-sphingomyelin solution in ethanol from step B2a into the 10 mL of 5µM fatty acid-free BSA solution from step B2b, and mix properly on a vortex mixer to generate the working solution (5 μM BODIPY FL-sphingomyelin + 5 μM BSA). Store the working solution at -20 °C, with strict protection from light.
Note: The purpose of defatted BSA in the working solution is to remove all residual pools of fluorescent lipid analogs, which will remain on the surface of plasma membranes after internalization of membrane-inserted lipid analogs, by “back-exchange” effects (Kok et al., 1989; Abreu et al., 2003).
Stain mESCs with the working solution of BODIPY FL C5-sphingolipid analogs for confocal imaging analysis
Remove all ESGRO growth medium from the overnight cell culture dishes and wash adherent cells three times with HBSS/HEPES buffer at RT, to remove residual ESGRO growth medium and floating dead cells. Apply sufficient working solution (prepared from the previous step) into cell culture dishes to completely cover the entire area where cells grow and incubate cells at 4 °C for 30 min with strict protection from light. The negative control sample will be incubated with the HBSS/HEPES buffer instead of BODIPY FL C5-sphingolipid working solution in this step.
Note: During the washing step, PBS solution should be added against the side of the well instead of directly on the top of cells, so that cells will not be dislodged. The healthy mESCs are able to attach to gelatin-coated cell culture dishes. The majority of the cells dislodged during washing are dead or apoptotic cells and must be discarded.
After 30 min incubation at 4 °C, completely remove all working solution and wash cells three times with ice-cold ESGRO growth medium. After washing, add adequate fresh RT ESGRO growth medium to cover the cells, and incubate cells in an incubator at 37 °C, 95% humidity, and 5% CO2 for 30 min. Strictly protect the incubation from light.
Wash cells with fresh ESGRO growth medium at RT. Stained cells on glass bottom dishes will be directly examined and imaged with the Zeiss LSM 780 UV confocal microscope.
Note: Cells can also be counter-stained with other dyes, such as nuclei staining with DAPI or DRQA5 (Fan et al., 2021).
The dynamic of BODIPY FL-sphingomyelins in stained mESCs will be analyzed using a Zeiss LSM 780 UV inverted confocal microscope equipped with a cell culture chamber that provides appropriate cell growth conditions (e.g., 37 °C, 95% humidity, and 5% CO2). Time-lapse images of stained mESCs are acquired every 5 min for up to 12 h. An example of the dynamic of BODIPY FL-sphingomyelins in mESCs can be found in our original research article (Fan et al., 2021).
Analyze mESCs stained with BODIPY FL C5-sphingolipid analogs by FACS
After incubation at 37 °C, as described in step C2, wash cells with HBSS/HEPES buffer three times, to completely remove the ESGRO growth medium. Completely disassociate mESCs cells by adding an appropriate amount of 0.05% trypsin (approximately 300–500 µL for each well of the 6-well plate), then incubating at 37 °C for 3 min. Wash cells twice with 30 mL of HBSS/HEPES buffer in 50-mL Corning tubes to completely remove residual trypsin and harvest cells by centrifuging at 300 × g for 3 min.
Note: Extra amount of trypsin and extended digestion time can be applied for this step to complete dissociate large mESC colonies and prevent formation of cell clumps.
Discard the supernatant and resuspend the cell pellets in 1 mL of HBSS/HEPES buffer, then transfer the cells into 5-mL round bottom polystyrene test tubes for FACS analysis.
Note: Samples must be strictly protected from light in the two steps above.
The parameters setup for visualization and quantification analysis by BD LSRFortessa FACS are based on the spectral characteristics of the BODIPY FL-sphingolipid described in Table 1.
Table 1. Spectral characteristics of BODIPY FL-sphingolipid (recommended in the manual of Molecular Probes)
Label Absorption/Emission (nm) Optical Filter BODIPY FL 505/511 Omega/Chroma
XF26, XF115/71010,41012
Data analysis
Confocal imaging analysis: the concentration of BODIPY FL-sphingolipid in cellular membranes can be determined from the confocal images based on their spectral properties. Generally, when excited with λex = 488 nm, cell organelles or regions containing high levels of BODIPY FL-sphingolipid analogs fluoresce red (λem ≥ 590 nm), while cell organelles or regions containing low levels of BODIPY FL-sphingolipid analogs fluoresce green (λem = 520–590) (Figure 3). Therefore, the concentration of BODIPY FL-sphingolipid analogs in cellular membranes can be estimated by quantifying the ratio of green to red fluorescence from microscopic images (Pagano et al., 1991; Chen et al., 1997). This method has been previously applied to estimate the concentration of BODIPY FL-lipid analogs in membranes of various cell types (Pagano and Chen, 1998; Marks et al., 2008). Please note that, when cultured in defined serum free medium, BODIPY FL-sphingomyelin stained mESCs only emit green fluorescence, as shown in our original research article (Fan et al., 2021).
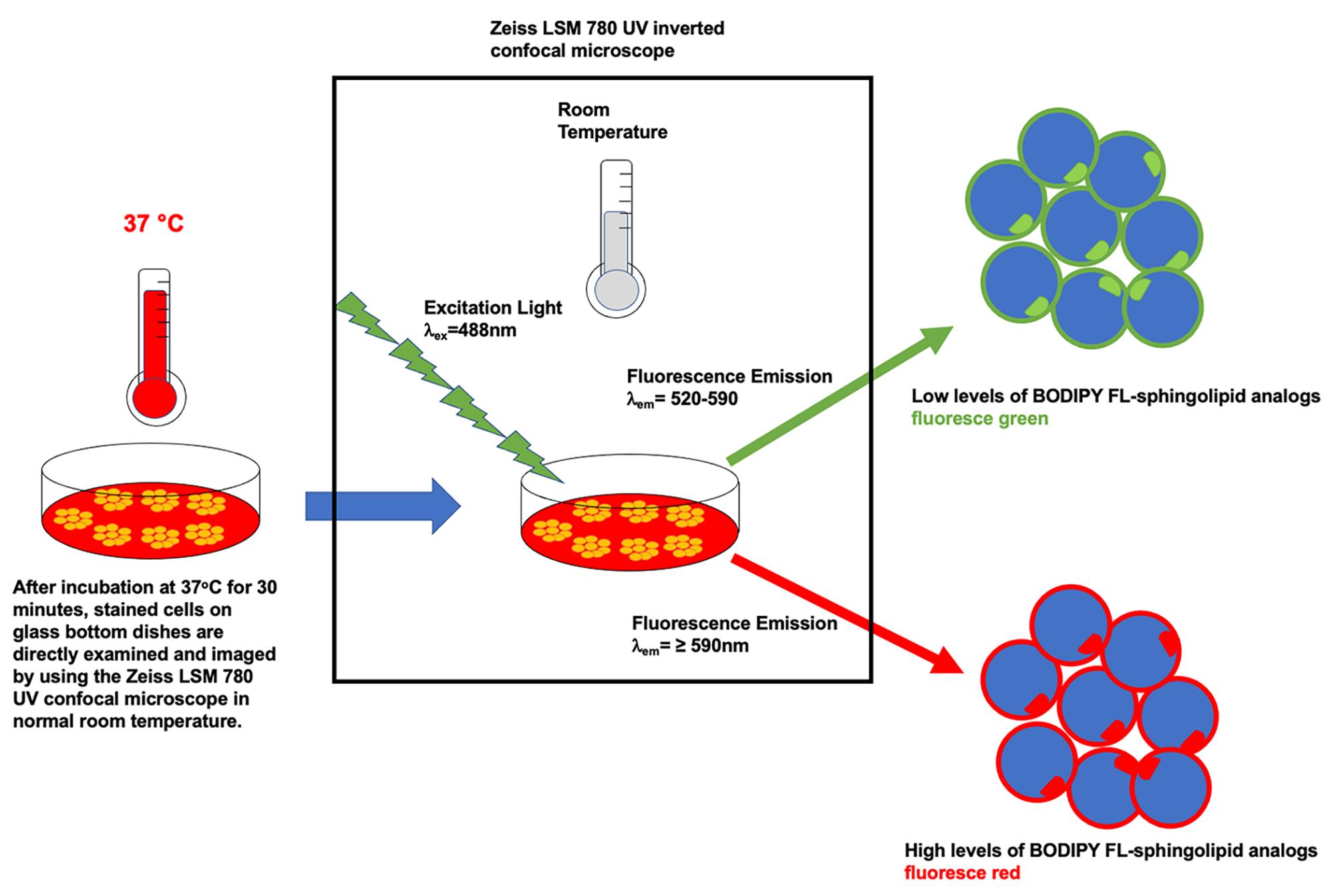
Figure 3. Schematic illustration of the image acquisition process using the Zeiss LSM 780 inverted confocal microscopeFACS data analysis: the FACS data from mESCs stained with BOPDIPY FL C5-sphingomyelin analogs are exported to BDFACSDivaTM software (version 8.0.1) for further analysis (Figure 4). All stained cells are initially gated by plotting forward scatter height (FSC-H) versus forward scatter area (FSC-A), to exclude doublets, then by plotting side scatter area (SSC-A) versus FSC-A to exclude the debris in the cell population. This gating strategy results in a population of “monodispersed state” single cells (Figure 4: the two plots on top of A and B). Next, the single parameter histogram of cell counts versus FSC-A is used to identify and gate the cell population carrying positive BODIPY fluorescence (FITC-A) after the comparison between positive sample (Figure 4B, the two middle plots) and negative control (Figure 4A, the two middle plots). Finally, the average FITC intensity of all counted single cells is calculated (Figure 4A and 4B, plots and tables in the bottom). The relative BODIPY fluorescence is calculated by normalizing the average intensity reading of each group against the wild-type (WT) group. Values are expressed as mean fluorescent intensity (MFI) ± standard error of mean from at least three independent experiments or biological replicates.
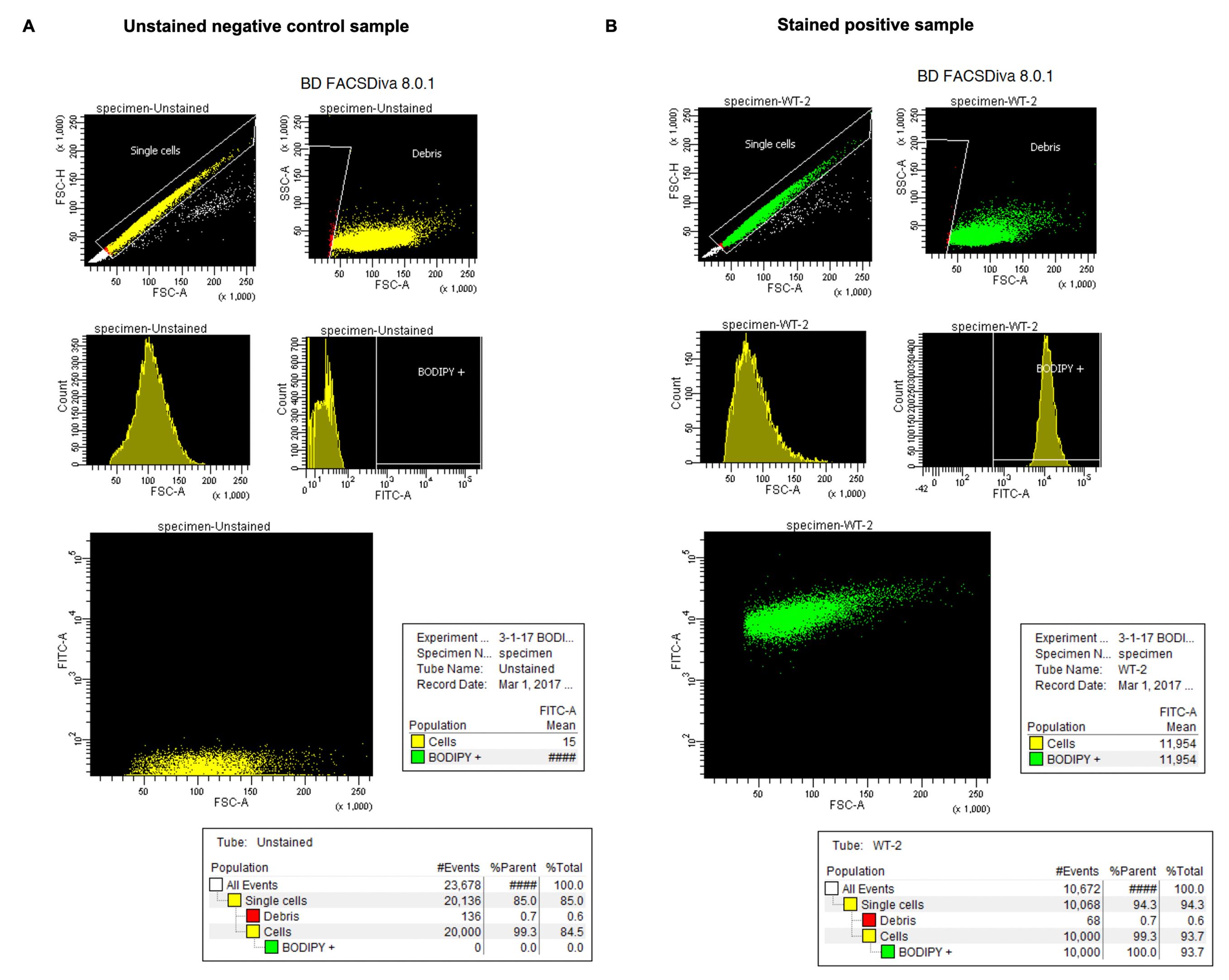
Figure 4. Determination of fluorescence intensity in mESCs stained with BODIPY FL C5-sphingomyelin by FACS. Data is analyzed with BDFACSDivaTM software version 8.0.1. for (A) unstained negative control and (B) stained positive mESCs.
Recipes
0.1% Gelatin solution
Reagent Final concentration Amount Gelatin (powder) 0.1%(W/V) 1 g H2O n/a 1,000 mL Total n/a 1,000 mL 70% ethanol
Reagent Final concentration Amount Ethanol (absolute) 70% 700 mL H2O n/a 300 mL Total n/a 1,000 mL Chloroform:ethanol (19:1 v/v)
Reagent Final concentration Amount Chloroform (absolute) 95% 0.95 mL Ethanol 5% 0.05 mL Total n/a 1 mL HBSS/HEPES buffer (pH 7.4)
Reagent Final concentration Amount HBSS 1× 495 mL HEPES buffer 10 mM 5 mL Total n/a 500 mL
Acknowledgments
We thank Dr. Carl Bortner and Mr. Jeff Tucker for critical reading of the manuscript. The authors also thank Yolanda L. Jones, National Institutes of Health Library, for editing assistance. The research related to this work was supported by the Intramural Research Program of National Institute of Environmental Health Sciences of the NIH Z01 ES102205 (to X. L.).
Fan, W., Tang, S., Fan, X., Fang, Y., Xu, X., Li, L., Xu, J., Li, J. L., Wang, Z. and Li, X. (2021). SIRT1 regulates sphingolipid metabolism and neural differentiation of mouse embryonic stem cells through c-Myc-SMPDL3B. Elife 10: e67452. doi: https://doi.org/10.7554/eLife.67452.
Competing interests
The authors declare that no competing interests exist.
References
- Abreu, M. S., Estronca, L. M., Moreno, M. J. and Vaz, W. L. (2003). Binding of a fluorescent lipid amphiphile to albumin and its transfer to lipid bilayer membranes. Biophys J 84(1): 386-399.
- Alessenko, A. V. and Albi, E. (2020). Exploring Sphingolipid Implications in Neurodegeneration. Front Neurol 11: 437.
- Bienias, K., Fiedorowicz, A., Sadowska, A., Prokopiuk, S. and Car, H. (2016). Regulation of sphingomyelin metabolism. Pharmacol Rep 68(3): 570-581.
- Chattopadhyay, A. and London, E. (1987). Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry 26(1): 39-45.
- Chen, C. S., Martin, O. C. and Pagano, R. E. (1997). Changes in the spectral properties of a plasma membrane lipid analog during the first seconds of endocytosis in living cells. Biophys J 72(1): 37-50.
- Fan, W., Tang, S., Fan, X., Fang, Y., Xu, X., Li, L., Xu, J., Li, J. L., Wang, Z. and Li, X. (2021). SIRT1 regulates sphingolipid metabolism and neural differentiation of mouse embryonic stem cells through c-Myc-SMPDL3B. Elife 10: e67452.
- Gault, C. R., Obeid, L. M. and Hannun, Y. A. (2010). An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol 688: 1-23.
- Grassi, S., Chiricozzi, E., Mauri, L., Sonnino, S. and Prinetti, A. (2019). Sphingolipids and neuronal degeneration in lysosomal storage disorders. J Neurochem 148(5): 600-611.
- Heinz, L. X., Baumann, C. L., Koberlin, M. S., Snijder, B., Gawish, R., Shui, G., Sharif, O., Aspalter, I. M., Muller, A. C., Kandasamy, R. K., et al. (2015). The Lipid-Modifying Enzyme SMPDL3B Negatively Regulates Innate Immunity. Cell Rep 11(12): 1919-1928.
- Hoekstra, D. and Kok, J. W. (1992). Trafficking of glycosphingolipids in eukaryotic cells; sorting and recycling of lipids. Biochim Biophys Acta 1113(3-4): 277-294.
- Johnson, I. D., Kang, H. C. and Haugland, R. P. (1991). Fluorescent membrane probes incorporating dipyrrometheneboron difluoride fluorophores. Anal Biochem 198(2): 228-237.
- Karolin, J., Johansson, L. B. A., Strandberg, L. and Ny, T. (1994). Fluorescence and Absorption Spectroscopic Properties of Dipyrrometheneboron Difluoride (Bodipy) Derivatives in Liquids, Lipid-Membranes, and Proteins. J Am Chem Soc 116(17): 7801-7806.
- Kok, J. W., Eskelinen, S., Hoekstra, K. and Hoekstra, D. (1989). Salvage of glucosylceramide by recycling after internalization along the pathway of receptor-mediated endocytosis. Proc Natl Acad Sci U S A 86(24): 9896-9900.
- Koval, M. and Pagano, R. E. (1989). Lipid recycling between the plasma membrane and intracellular compartments: transport and metabolism of fluorescent sphingomyelin analogues in cultured fibroblasts. J Cell Biol 108(6): 2169-2181.
- Koval, M. and Pagano, R. E. (1990). Sorting of an internalized plasma membrane lipid between recycling and degradative pathways in normal and Niemann-Pick, type A fibroblasts. J Cell Biol 111(2): 429-442.
- Koval, M. and Pagano, R. E. (1991). Intracellular transport and metabolism of sphingomyelin. Biochim Biophys Acta 1082(2): 113-125.
- Laude, A. J. and Prior, I. A. (2004). Plasma membrane microdomains: organization, function and trafficking. Mol Membr Biol 21(3): 193-205.
- Marks, D. L., Bittman, R. and Pagano, R. E. (2008). Use of Bodipy-labeled sphingolipid and cholesterol analogs to examine membrane microdomains in cells. Histochem Cell Biol 130(5): 819-832.
- Martin, O. C. and Pagano, R. E. (1987). Transbilayer movement of fluorescent analogs of phosphatidylserine and phosphatidylethanolamine at the plasma membrane of cultured cells. Evidence for a protein-mediated and ATP-dependent process(es). J Biol Chem 262(12): 5890-5898.
- Martin, O. C. and Pagano, R. E. (1994). Internalization and sorting of a fluorescent analogue of glucosylceramide to the Golgi apparatus of human skin fibroblasts: utilization of endocytic and nonendocytic transport mechanisms. J Cell Biol 125(4): 769-781.
- Mayor, S., Presley, J. F. and Maxfield, F. R. (1993). Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J Cell Biol 121(6): 1257-1269.
- Pagano, R. E. (1990). The Golgi apparatus: insights from lipid biochemistry. Biochem Soc Trans 18(3): 361-366.
- Pagano, R. E. and Chen, C. S. (1998). Use of BODIPY-labeled sphingolipids to study membrane traffic along the endocytic pathway. Ann Ny Acad Sci 845: 152-160.
- Pagano, R. E., Martin, O. C., Kang, H. C. and Haugland, R. P. (1991). A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol 113(6): 1267-1279.
- Pagano, R. E., Watanabe, R., Wheatley, C. and Dominguez, M. (2000). Applications of BODIPY-sphingolipid analogs to study lipid traffic and metabolism in cells. Methods Enzymol 312: 523-534.
- Rosenwald, A. G. and Pagano, R. E. (1993). Intracellular transport of ceramide and its metabolites at the Golgi complex: insights from short-chain analogs. Adv Lipid Res 26: 101-118.
- Schnaar, R. L. and Kinoshita, T. (2015). Glycosphingolipids. (3rd edition). In: Varki, A., Cummings, R. D., Esko, J. D., Stanley, P., Hart, G. W., Aebi, M., Darvill, A. G., Kinoshita, T., Packer, N. H., Prestegard, J. H., et al. (Eds.) Essentials of Glycobiology. 125-135.
- Tanosaki, S., Tohyama, S., Kishino, Y., Fujita, J. and Fukuda, K. (2021). Metabolism of human pluripotent stem cells and differentiated cells for regenerative therapy: a focus on cardiomyocytes. Inflamm Regen 41(1): 5.
- Wolf, D. E., Winiski, A. P., Ting, A. E., Bocian, K. M. and Pagano, R. E. (1992). Determination of the transbilayer distribution of fluorescent lipid analogues by nonradiative fluorescence resonance energy transfer. Biochemistry 31(11): 2865-2873.
Article Information
Copyright
Fan and Li. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Fan, W. and Li, X. (2022). Using BODIPY FL-Sphingolipid Analogs to Study Sphingolipid Metabolism in Mouse Embryonic Stem Cells. Bio-protocol 12(22): e4555. DOI: 10.21769/BioProtoc.4555.
- Fan, W., Tang, S., Fan, X., Fang, Y., Xu, X., Li, L., Xu, J., Li, J. L., Wang, Z. and Li, X. (2021). SIRT1 regulates sphingolipid metabolism and neural differentiation of mouse embryonic stem cells through c-Myc-SMPDL3B. Elife 10: e67452.
Category
Stem Cell > Embryonic stem cell > Cell staining
Cell Biology > Cell imaging > Confocal microscopy
Cell Biology > Cell staining > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









