- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Collection of Xylem Exudates from the Model Plant Arabidopsis and the Crop Plant Soybean
Published: Vol 12, Iss 19, Oct 5, 2022 DOI: 10.21769/BioProtoc.4520 Views: 2744
Reviewed by: Ashish RanjanPooja VermaMalgorzata LichockaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2855 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1975 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1763 Views
Abstract
A number of molecules, such as secreted peptides, have been shown to mediate root-to-shoot signaling in response to various conditions. The xylem is a pathway for water and molecules that are translocated from roots to shoots. Therefore, collecting and analyzing xylem exudates is an efficient approach to study root-to-shoot long-distance signaling. Here, we describe a step-by-step protocol for the collection of xylem exudate from the model plant Arabidopsis and the crop plant soybean (Glycine max). In this protocol, we can collect xylem exudate from plants cultured under normal growth conditions without using special equipment.
Graphical abstract:

Xylem exudates on the cut surfaces of an Arabidopsis hypocotyl and a soybean internode.
Background
Plants consist of multiple organs that play distinctive roles during plant life. Hence, organ-to-organ communication is indispensable for plants to adapt to environmental stresses and maintain homeostasis at the whole-plant level. It is known that various types of macromolecules, such as phytohormones, RNAs, proteins, and peptides mediate long-distance signaling (Okamoto et al., 2015, 2016; Lu et al., 2020). The xylem plays an important role in the translocation of water and micronutrients from roots to shoots. In addition, long-distance signaling molecules, including small secreted peptides and phytohormones, are translocated from roots to shoots through the xylem. Therefore, collecting and analyzing xylem exudates is an effective approach to identify novel long-distance mobile molecules.
To date, several methods have been developed to collect xylem exudates (Buhtz, 2004, Goodger et al., 2005; Dafoe and Constabel, 2009), and the appropriateness of a given method is dependent on plant species and conditions. One method uses a pressure chamber to obtain xylem exudate from woody plants. In this method, roots are placed inside a sealed chamber, and pressurized gas is added to the chamber. This method can be applied to woody plants that have mechanically stronger tissues than herbaceous plants, and a precise protocol was written by Flajšman et al. (2017). Another method depends on root pressure and is usually applied to herbaceous plants whose tissues are soft. Root pressure is one of the driving forces of xylem sap. Accumulation of solutes in root xylem results in low water potential and generates positive hydrostatic pressure in the xylem (Taiz et al., 2015). Although the quantity of xylem exudates obtained with this method depends on plant species and conditions, this method requires no special equipment and has been applied to many herbaceous plants, including Arabidopsis thaliana (Iwai et al., 2003; Buhtz et al., 2004; Kehr et al., 2005; Alvarez et al., 2006; Djordjevic et al., 2007; Ligat et al., 2011; Ko et al., 2014; Tabata et al., 2014; Okamoto et al., 2015; Luo and Zhang, 2019). However, the collection of xylem exudates is likely not familiar to many researchers, and it is difficult to find a step-by-step protocol for this method.
Previously, we collected xylem exudate from soybean to conduct peptidome analysis and identified various endogenous root-to-shoot mobile peptides (Okamoto et al., 2015). In addition, we analyzed xylem exudate from the model plant Arabidopsis thaliana and detected mature CLAVATA3/ESR-related 2 peptides (Okamoto et al., 2022a). In this protocol, we describe step-by-step procedures for the collection of xylem exudates from both the model plant Arabidopsis and the crop plant soybean.
Part I. Collection of xylem exudates from Arabidopsis
Materials and Reagents
1.5 mL tube (BIO-BIK, catalog number: ST-0150F)
Plastic square dish (140 mm × 100 mm, height 18 mm/EIKEN, catalog number: AW2000)
Plastic Petri dish (ø 85 mm, height 20 mm/EIKEN, catalog number: AP2310)
Razor blade (FEATHER, FA-10)
Micropipette [Nichiryo, catalog number: 00-NPX2-2 (0.1–2 µL)]
Arabidopsis seeds
Ethanol (FUJIFILM Wako, catalog number: 057-00451)
Tween 20 (MP Biomedicals, catalog number: 194841)
Sodium hypochlorite solution (FUJIFILM Wako, catalog number: 197-02206)
Murashige and Skoog (MS) plant salt mixture (FUJIFILM Wako, catalog number: 392-00591)
Gamborg’s vitamin solution 1,000× (Sigma-Aldrich, catalog number: G1019)
Sucrose (FUJIFILM Wako, catalog number: 196-00015)
Agar (FUJIFILM Wako, catalog number: 016-11875)
Culture media (100 mL) (see Recipes)
Equipment
Growth chamber (Nippon Medical & Chemical Instruments, model: LH-241S)
Procedure
Place Arabidopsis seeds in 1.5 mL tubes.
Add 1 mL of 70% ethanol and wash the seeds for 1 min.
Remove the ethanol solution.
Add 1 mL of H2O, 10 µL of 10% Tween 20, and 50 µL of sodium hypochlorite solution and mix using a vortex mixer.
Wash the seeds for 15 min by gently inverting the tubes.
Remove the solution and rinse the seeds five times with sterilized water.
Sow the seeds in plastic Petri dishes with culture media (see Recipes).
Incubate at 4 °C for three days.
Cover the dishes with aluminum foil to grow seedlings in darkness and incubate at 23 °C for five days.
Remove the aluminum foil. At this stage, the seedlings are etiolated with elongated hypocotyls (Figure 1A). Note that further darkness affects seedling viability.
Grow the seedlings under a 10 h light/14 h dark cycle at 23 °C for five days. The cotyledons will become green (Figure 1B).
Transfer the Arabidopsis seedlings to plastic square dishes with culture media [with or without 1% sucrose (see Notes 1)] on a clean bench.
Place the dishes vertically and incubate the plants under a 10 h light [100 µmol/m2s photosynthetic photon flux density (PPFD)]/14 h dark cycle at 23 °C for 14 days (Figure 1C).
Cut the hypocotyls with a razor blade (Figure 1D). Within 20–40 min, xylem exudates will form droplets on the cut surface of the cotyledon (Figure 1E).
Wipe the first droplets with soft paper to avoid contamination of phloem sap and then collect the droplets with a micropipette. The droplets can be sampled at approximately 30 min intervals.
Store the collected exudates at -80 °C.
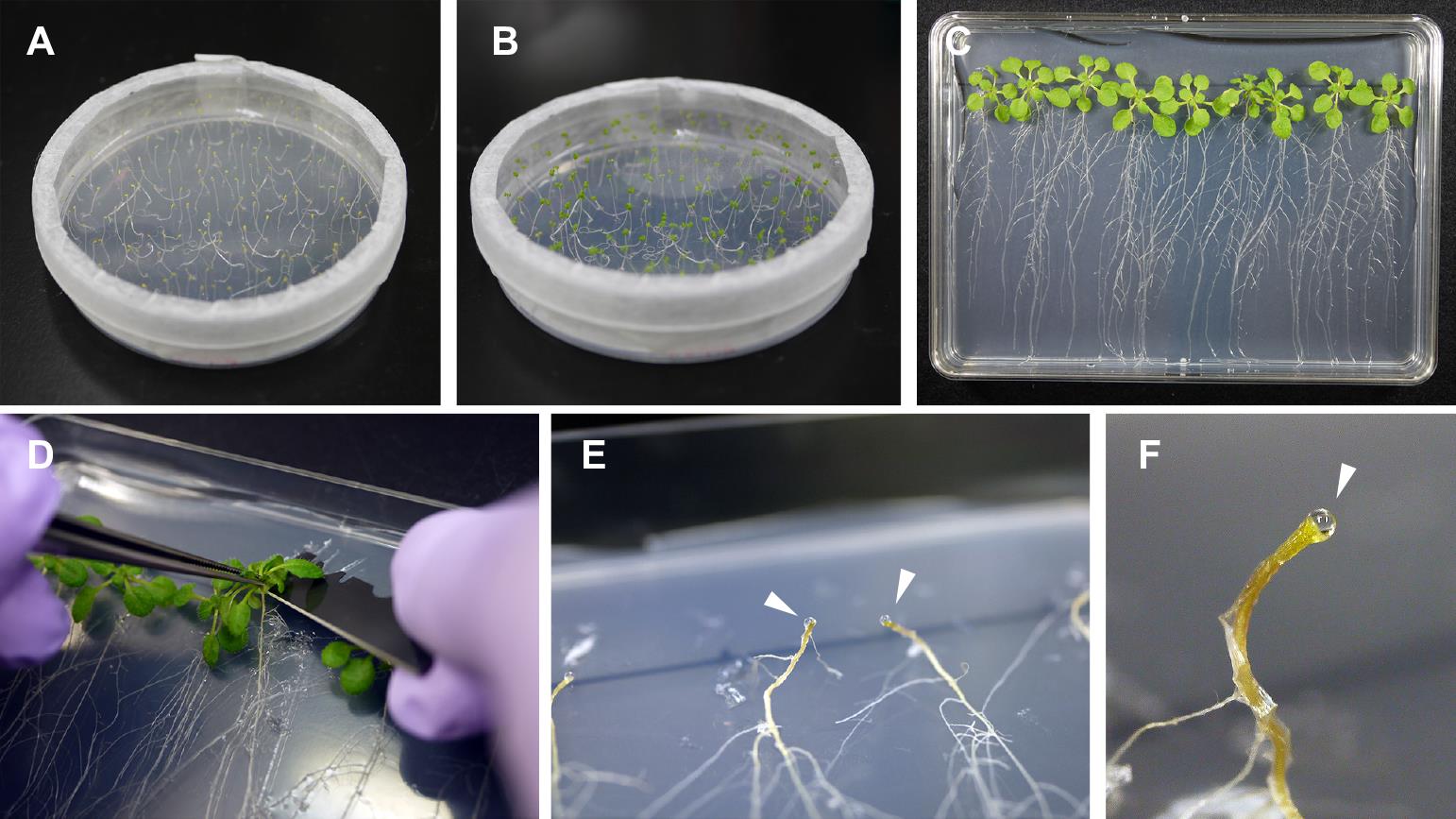
Figure 1. Collection of xylem exudates from Arabidopsis. (A) Arabidopsis seedlings at five days after germination (DAG), grown under dark conditions. (B) Arabidopsis seedlings at 10 DAG, incubated under a light/dark cycle for five days. (C) Arabidopsis plants at 24 DAG. (D) Arabidopsis hypocotyls are cut with a razor blade. (E) and (F) Xylem exudates form droplets on the cut surface of hypocotyls, and these droplets are collected with a micropipette. Each droplet is indicated by a triangle.
Notes
In our study (Okamoto et al., 2022a), because the target peptide gene responds to low-carbon conditions, Arabidopsis was cultured without sucrose from 10–24 DAG. The exudate was collected for 4 h, and approximately 320 µL of exudate was obtained from 360 plants. Application of sucrose to the culture medium increases the yield of xylem exudates.
The amount of xylem exudates obtained from Arabidopsis is too small to conduct proteomics analysis (approximately 1 µL/plant), so the purpose of analyzing Arabidopsis xylem exudate should be the detection of interesting (target) molecule(s).
To conduct a comprehensive analysis, a larger amount of xylem exudates must be collected from larger plants (see Part II).
Recipes
Culture media (100 mL)
Reagent Final concentration Amount Murashige and Skoog (MS) salt mixture 1/2× 0.23 g Sucrose 1 or 0% (w/v) 1 or 0 g Gamborg’s vitamin solution 1,000× 1× 100 µL Adjust the pH to 5.7 and add 0.8 g agar; then, autoclave the solution for 20 min at 121 °C.
Part II. Collection of xylem exudates from soybean
Materials and Reagents
Soybean seeds
50 mL centrifuge tubes (Labcon, catalog number: 3181-345)
Plastic square dish (140 mm × 100 mm, height 18 mm/EIKEN, catalog number: AW2000)
Silicon tube [select a tube whose inner diameter is fitted to or slightly larger than (less than 1 mm larger) the cut stems. In the case of soybean, the diameter of the lower internodes is 4–7 mm approximately seven weeks after germination. The length of the tube depends on the length of the flat needle or micropipette tip. When the flat needle described below was used, we cut the tube into approximately 8 cm lengths [AsOne, catalog number: 6-586-12 (4 mm i.d.), 6-586-17 (5 mm i.d.), 6-586-20 (6 mm i.d.), and 6-586-25 (7 mm i.d.)]
Plastic pot (ø 150 mm, height 127 mm)
Garden shears
Flat needle (AsOne, catalog number: 1-2752-01)
5 mL syringe (TERUMO, catalog number: SS-05SZ)
Ethanol (FUJIFILM Wako, catalog number: 057-00451)
Vermiculite
Clay
Nutrition solution for soybean (1 L) (see Recipes)
Equipment
Greenhouse or growth chamber (Nippon Medical & Chemical Instruments, model: LH-411PFQDT-SP)
Centrifuge (Kubota, model: 2410)
Procedure
Place seeds (less than 25 mL to facilitate washes) in 50 mL centrifuge tubes and add approximately 40 mL of 70% ethanol to wash the seeds by inverting gently for 1 min.
Rinse the seeds with distilled water five times.
Place soft wet paper on plastic square dishes and arrange the seeds on the paper (approximately 25 seeds/dish) (Figure 2A).
Cover the seeds with wet, soft paper and place a lid on the dishes to maintain higher humidity (Figure 2B).
Incubate the seeds in the square dishes under a 16 h light (200 µmol/m2s PPFD)/8 h dark cycle at 23 °C for two days.
Select healthy seedlings (e.g., relatively long roots, greening cotyledon) and transplant them into vermiculite in plastic pots.
Place the pots in a growth chamber [16 h light (900 µmol/m2s PPFD)/8 h dark, 23 °C, and humidity not controlled] or greenhouse and supply a sufficient amount of nutrition solution. The frequency of solution application depends on the plant age, conditions, and weather (in the case of a greenhouse).
Approximately seven weeks after germination, cut the hypocotyls or lower internodes (first or second internodes from hypocotyls) with garden shears (Figure 2C, D). The hypocotyls and the lower internodes are too hard to cut with a razor blade.
Wash the cut surface with distilled water by dripping water on the surface.
Allow the cut stem to leak exudates for 20 min to avoid contamination from phloem sap.
Wipe the cut surface and the side of the stem with soft paper and wrap the stem just beneath the cut surface with clay (make a clay ring around the stem) (Figure 2E). Note that the droplets on the side of the stem prevent clay from sticking to the stem; therefore, the droplets need to be completely wiped off.
Place and press a silicon tube into the clay to avoid leakage of xylem exudates (Figure 2F). The gap between the stem and tube should be sealed with clay.
Collect xylem exudates using a syringe with a flat needle or micropipette. The exudates can be sampled at 20–30 min intervals.
Clay particles sometimes contaminate the exudates. To remove them from the xylem exudates, centrifuge at 2,600 × g for 10 min (and optimally at 4 °C) and transfer the supernatant to a new tube.
Store the xylem exudates at -80 °C.

Figure 2. Collection of xylem exudates from soybean. (A) Soybean seeds were arranged on soft wet paper in a plastic square dish. (B) The seeds were covered with wet paper, and a lid was placed on the dishes. (C) Lower internodes (1st or 2nd internodes from the bottom) of soybean. The plants were grown in a growth chamber (PPFD = 900 µmol/m2s). (D) Hypocotyl or lower internodes were cut with garden shears. (E) The stem just beneath the cut surface was wrapped with clay. (F) A silicon tube is pressed onto the cut stem, and xylem exudates accumulate in the tube. (G) From cutting the internodes to the fitting of a tube (C–F), it takes less than 1 min per plant, so this method is applicable to collecting xylem exudates from a large number of plants for comprehensive analysis.
Notes
A larger quantity of exudates will be obtained when plants are cultured in a greenhouse rather than in a growth chamber.
Under greenhouse conditions, approximately 60 mL of xylem exudate was obtained from 12 soybean plants (45 DAG) during the five hours after cutting the stems (approximately 5 mL/plant).
The yield of xylem exudates can decrease under stressed conditions, such as low-N and salt stress conditions. In our case, when soybean plants were cultured under a low-N (1.2 mM NH4NO3) condition for the last five days, approximately 20 mL of xylem exudate was obtained from 11 soybean plants in five hours. When soybean plants were cultured with the nutrition solution containing 50 mM NaCl for the last five days, no xylem exudates were obtained.
This method can be applied to various dicot plants. Under greenhouse conditions, approximately 120 mL of xylem exudate was obtained from 14 tomato (Solanum lycopersicum) plants (45 DAG) during the five hours after cutting the stems (approximately 8.6 mL/plant), and seven endogenous peptides were identified (Okamoto et al., 2022b).
Recipes
Nutrition solution for soybean (1 L)
Reagent Final concentration Amount NH4NO3 6.0 mM 480 mg CaCl2·2H2O 1.8 mM 262 mg MgSO4·7H2O 1.0 mM 245 mg KH2PO4 0.8 mM 108.8 mg K2SO4 0.28 mM 49.6 mg Fe(III)-EDTA 0.1 mM 43.9 mg MnSO4·5H2O 5.9 µM 1.43 mg H3BO3 4.0 µM 0.25 mg CuSO4·5H2O 1.0 µM 0.25 mg ZnSO4·7H2O 0.87 µM 0.25 mg Na2MoO4·2H2O 0.21 µM 0.05 mg CoCl2·6H2O 0.13 µM 0.03 mg The reagents are dissolved in desalted water, and total volume is adjusted to 1 L.
The pH is adjusted to 6.0.
Acknowledgments
These procedures are based on Okamoto et al. (2022a) and Okamoto et al. (2015). This work was supported by JST PRESTO (JPMJPR17Q2 to S.O.) and the MEXT Leading Initiative for Excellent Young Researchers (to S.O.).
Competing interests
The authors declare no competing interests.
References
- Alvarez, S., Goodger, J. Q. D., Marsh, E. L., Chen, S., Asirvatham, V. S., and Schachtman, D. P. (2006). Characterization of the Maize Xylem Sap Proteome. J Proteome Res 5(4): 963-972.
- Buhtz, A., Kolasa, A., Arlt, K., Walz, C., and Kehr, J. (2004). Xylem sap protein composition is conserved among different plant species. Planta 219(4): 610-618.
- Dafoe, N. J., and Constabel, C. P. (2009). Proteomic analysis of hybrid poplar xylem sap. Phytochemistry 70(7): 856-863.
- Djordjevic, M. A., Oakes, M., Li, D. X., Hwang, C. H., Hocart, C. H., and Gresshoff, P. M. (2007). The Glycine max Xylem Sap and Apoplast Proteome. J Proteome Res 6(9): 3771-3779.
- Flajšman, M., Mandelc, S., Radišek, S., and Javornik, B. (2017). Xylem Sap Extraction Method from Hop Plants. Bio-protocol 7(6): e2172.
- Goodger, J. Q. D., Sharp, R. E., Marsh, E. L., Schachtman D. P. (2005). Relationships between xylem sap constituents and leaf conductance of well-watered and water-stressed maize across three xylem sap sampling techniques. J Exp Bot 56(419): 2389-2400.
- Iwai, H., Usui, M., Hoshino, H., Kamada, H., Matsunaga, T., Kakegawa, K., Ishii, T., and Satoh, S. (2003). Analysis of Sugars in Squash Xylem Sap. Plant Cell Physiol 44(6): 582-587.
- Kehr, J., Buhtz, A., and Giavalisco, P. (2005). Analysis of xylem sap proteins from Brassica napus. BMC Plant Biol 5: 11.
- Ko, D., Kang, J., Kiba, T., Park, J., Kojima, M., Do, J., Kim, K. Y., Kwon, M., Endler, A., Song, W. Y., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc Natl Acad Sci USA 111(19): 7150-7155.
- Ligat, L., Lauber, E., Albenne, C., San Clemente, H., Valot, B., Zivy, M., Pont-Lezica, R., Arlat, M., and Jamet, E. (2011). Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11(9): 1798-1813.
- Lu, X., Liu, W., Wang, T., Zhang, J., Li, X., and Zhang, W. (2020). Systemic Long-Distance Signaling and Communication Between Rootstock and Scion in Grafted Vegetables. Frontiers in Plant Science 11: 460.
- Luo, J. S., and Zhang, Z. (2019). Proteomic changes in the xylem sap of Brassica napus under cadmium stress and functional validation. BMC Plant Biology 19: 280.
- Okamoto, S., Suzuki, T., Kawaguchi, M., Higashiyama, T., and Matsubayashi, Y. (2015). A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. Plant J 84: 611-620.
- Okamoto, S., Tabata, R., and Matsubayashi, Y. (2016). Long-distance peptide signaling essential for nutrient homeostasis in plants. Curr Opin Plant Biol 34: 35-40.
- Okamoto, S., Kawasaki, A., Makino, Y., Ishida, T., and Sawa, S. (2022a). Long-distance translocation of CLAVATA3/ESR-related 2 peptide and its positive effect on root sucrose status. Plant Physiol 89(4): 2357-2367.
- Okamoto, S., Kawasaki, A., and Makino, Y. (2022b). Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates. Life 12: 592.
- Tabata, R., Sumida, K., Yoshii, T., Ohyama, K., Shinohara, H., and Matsubayashi, Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346(6207): 343-346.
- Taiz, L., Zeiger, E., Moller I. M. and Murphy A. (2015). Plant Physiology and Development, Sixth Edition. Sinauer Associates.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
OKamoto, S. and Kawasaki, A. (2022). Collection of Xylem Exudates from the Model Plant Arabidopsis and the Crop Plant Soybean. Bio-protocol 12(19): e4520. DOI: 10.21769/BioProtoc.4520.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









