- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorescent Labeling of Small Extracellular Vesicles (EVs) Isolated from Conditioned Media
Published: Vol 12, Iss 12, Jun 20, 2022 DOI: 10.21769/BioProtoc.4447 Views: 5671
Reviewed by: Alba BlesaSeda EkiciAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Generation of Giant Unilamellar Vesicles (GUVs) Using Polyacrylamide Gels
Eric Parigoris [...] Unai Silvan
Nov 5, 2020 4820 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1441 Views

Production of Genetically Engineered Extracellular Vesicles for Targeted Protein Delivery
Leyla A. Ovchinnikova [...] Yakov A. Lomakin
Nov 5, 2025 1628 Views
Abstract
Extracellular vesicles (EVs), such as exosomes, are produced by all known eukaryotic cells, and constitute essential means of intercellular communication. Recent studies have unraveled the important roles of EVs in migrating to specific sites and cells. Functional studies of EVs using in vivo and in vitro systems require tracking these organelles using fluorescent dyes or, alternatively, transfected and fluorescent-tagged proteins, located either intravesicularly or anchored to the EV bilayer membrane. Due to design simplicity, the fluorescent dye might be a preferred method if the cells are difficult to modify by transfection or when the genetic alteration of the mother cells is not desired. This protocol describes techniques to label cultured cell-derived EVs, using lipophilic DiR [DiIC18(7) (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindotricarbocyanine Iodide)] fluorophore. This technique can be used to study the cellular uptake and intracellular localization of EVs, and their biodistribution in vivo, which are crucial evaluations of any isolated EVs.
Keywords: Extracellular vesiclesBackground
There are several subtypes of extracellular vesicles (EVs), and among them, exosomes are one of the smallest ones, with a size range between 30 nm and 120 nm (reviewed in Théry et al., 2002). Exosomes contain signature molecules, such as tetraspanins (CD9, CD63, and CD81), and are characterized by specific biogenesis originating in the endosomal pathway (reviewed in Huotari and Helenius, 2011). The EVs can carry and transfer various cargo to other cells, including proteins, metabolites, miRNA, and lipids. One of the exciting research areas is the affinity of exosomes to specific tissues and regions, depending on the characteristics of these EVs. Hence, the biodistribution of EVs should be studied, and EV imaging is required to accomplish this goal. Several methods can be used to track EVs in animal cells and tissues. For instance, bioluminescence imaging (BLI) (Gangadaran et al., 2017) is one of the methods with the highest signal-to-noise ratio, but it suffers from low temporal resolution. Magnetic resonance imaging (MRI) (Busato et al., 2016, 2017) can also be used, but this method is associated with low sensitivity and high operation cost. Finally, fluorescence-based imaging of EVs provides a sensitive method with the highest spatial resolution (Corso et al., 2019). Among the fluorescence-based techniques, GFP protein can be expressed with the lowest penetration, which does not allow for noninvasive in vivo imaging, but has high resolution. Near-infrared (NIR) fluorescence imaging using lipophilic dyes, such as DiR [DiIC18(7) (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindotricarbocyanine Iodide)], can also be used. The two long 18-carbon chains insert into the vesicle membrane avidly, resulting in a negligible dye transfer between EVs. The near IR fluorescent lipophilic carbocyanine DiOC18(7) ('DiR') is weakly fluorescent in water but highly fluorescent and quite photostable when incorporated into membranes. The sulfonate residues incorporated into this DiI analog improve water solubility. Moreover, DiI has an extremely high extinction coefficient and short excited-state lifetimes (~1 nanosecond) in the lipid-rich environment. The DiR shows excitation/emission at 710/760 nm, which, along with its NIR properties, makes this an ideal fluorophore for in vivo EV imaging studies, given the significantly reduced auto-fluorescence from the animal at higher wavelengths (Cook et al., 2015; Somanchi, 2016; Zhao et al., 2021). While there is no perfect method, DiR–based labeling can provide a flexible and low-cost alternative to other methods used for systematic studies of exosomes. This paper describes a technique that uses a lipophilic DiR dye to stain EVs obtained from cell culture, such as cultured macrophages. This approach can be used to track the vesicles for in vitro uptake and in vivo biodistribution studies in animal models.
Materials and Reagents
Polypropylene open-top ultracentrifuge tubes (Beckman Coulter, catalog number: 326823)
Nalgene Rapid-Flow Sterile Single Use Vacuum 0.2 µm Filter 500 mL Unit (Thermo Fisher, catalog number: 566-0020)
Amicon Ultra Centrifugal Filter Ultracell 10,000 MWCO (Millipore Sigma, catalog number: UFC801024)
Syringe Filters, Polyethersulfone (PES) membrane, 0.22 µm (GenClone, catalog number: 25-244)
Greiner Bio-One CellStar μClear 96-Well, Cell Culture-Treated, Flat-Bottom Microplate (Greiner Bio-One, catalog number: 655090)
DiR'; DiIC18(7) (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindotricarbocyanine Iodide) (Thermo Fisher Scientific, catalog number: D12731)
Pierce Protease Inhibitor Tablets (Thermo Fisher Scientific, catalog number: A32963)
1× Dulbecco’s Phosphate Buffered Saline (DPBS) with Calcium and Magnesium (Thermo Fisher Scientific, catalog number: 14040117)
OptiMEM I Reduced Serum Medium, no phenol red (Thermo Fisher, catalog number: 11058021)
Phosphate Buffered Saline (PBS, 1×), sterile filtered (Thermo Fisher, catalog number: J61196.AP)
DAPI solution (1 mg/mL, Thermo Fisher, catalog number: 62248)
Pierce Protease Inhibitor Tablets, EDTA-Free (Thermo Fisher, catalog number: A32955) (See Recipes)
MicroBCA Protein Assay Kit (Thermo Fisher, catalog number: 23235)
Media containing exosome-depleted fetal bovine serum (FBS) and penicillin/streptomycin (see Recipes)
Control Sample for NanoSight quantification (see Recipes)
Protease inhibitor cocktail (PI) from Pierce Protease Inhibitor Tablets, EDTA-Free (see Recipes)
Equipment
Optima XPN-90 IVD ultracentrifuge (Beckman, catalog number: B10052) with SW 32 Ti swinging-bucket rotor (Beckman, catalog number: 369650)
Sorvall Legend Micro 21R Microcentrifuge (Thermo Scientific, catalog number: 75002447)
Cytation 5 multi-mode reader (Agilent, catalog number: BTCYT5V)
NanoSight LM10 (Malvern Panalytical, catalog number: EOS3085)
Software
GraphPad Prism 9 (GraphPad Software)
Gen5 Software (Agilent)
Procedure
The overall scheme for the EV purification and labeling with DiR dye is presented in Figure 1.
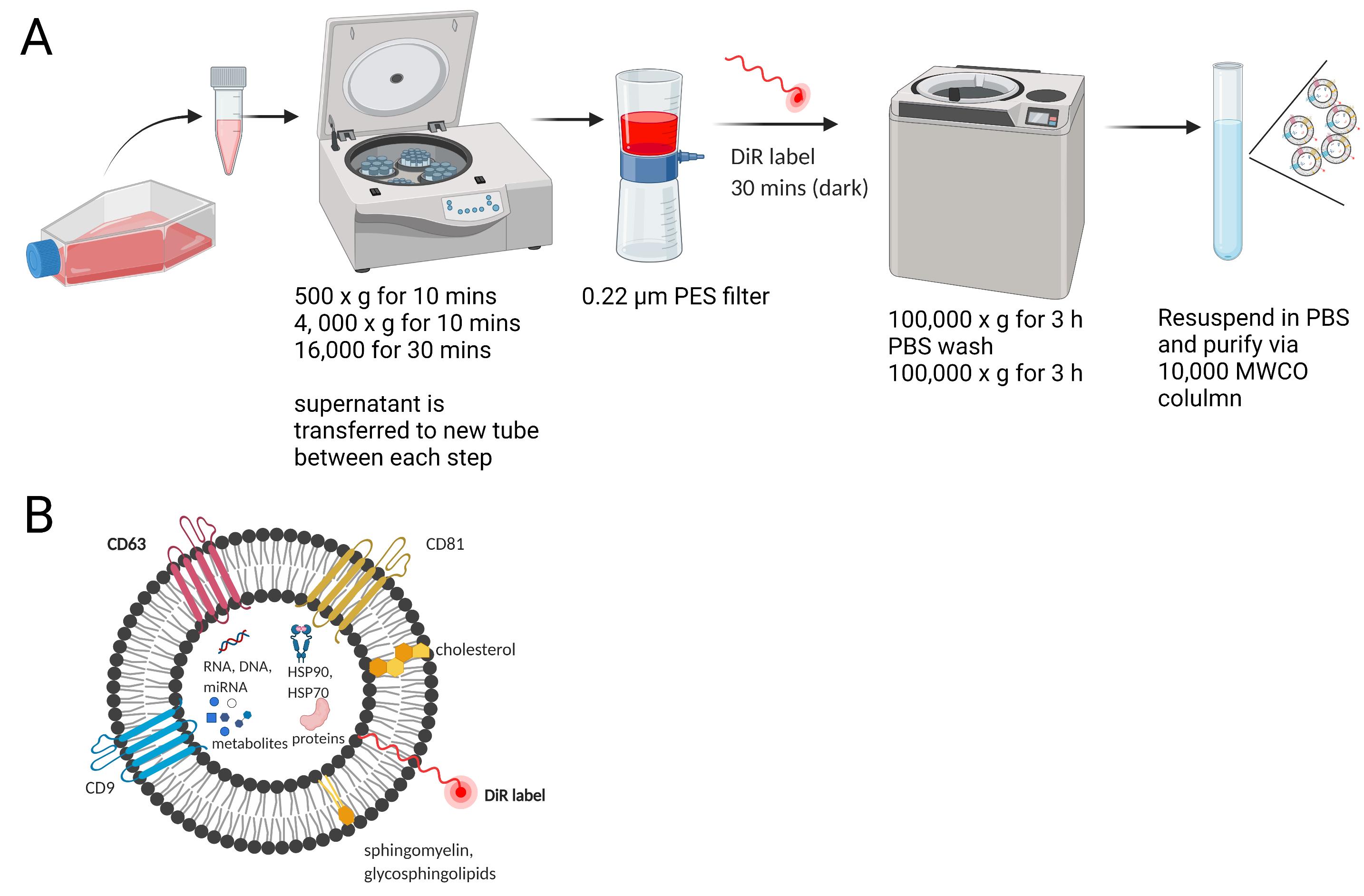
Figure 1. Experimental setup for the EV purification and labeling with DiR. A. The EV purification and labeling scheme. B. The EV is labeled with a DiR or a similar fluorogenic dye.
Collection of the cell culture conditioned media
Set the ultracentrifuge and benchtop refrigerated centrifuge to 4°C, approximately 30 min before collecting the supernatant. Prepare a bucket with ice before collecting the cell culture media (conditioned cell culture media; see Recipes for an example of media).
Notes:
Use sterile reagents and proper aseptic technique at all steps to avoid contamination.
Before loading the tubes, it is recommended to refrigerate the rotor and the tubes.
Collect 15 mL of conditioned cell culture media at the desired time (such as 24 h). Add the protease inhibitor cocktail (PI) at 1% (v/v) to the conditioned media.
Centrifuge media at 500 × g and 4°C for 10 min, to remove cells. Carefully decant supernatant to sterile centrifuge tubes—a 25 mL serological pipette can be used.
Centrifuge supernatant at 4,000 × g and 4°C for 10 min. Carefully decant supernatant to new sterile centrifuge tubes.
Centrifuge supernatant at 16,000 × g and 4°C for 30 min. Carefully decant supernatant to new sterile centrifuge tubes.
Filter-sterilize the supernatant through a 0.22 µm PES filter into a new sterile centrifuge tube.
DiR labeling
Prepare DiIC18(7) (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindotricarbocyanine Iodide) (DiR) stock at 2.5 mg/mL (2.47 mM) in ethanol.
Add the appropriate volume of DiR stock to the filtered supernatant from the steps above, to reach a 5 µM final concentration. Mix by pipetting.
Incubate DiR with the filtered supernatant in the dark at room temperature (RT) for 30 min, with gentle inversion every 5 minutes.
Purification of small EVs
Sterilize items for ultracentrifugation by UV light exposure of buckets, centrifuge tubes, and forceps (for removing ultracentrifuge tubes from buckets/rotor) for at least 30 min.
Fill the ultracentrifuge tubes to a volume of approximately 37 mL.
Note: When there is not enough supernatant to fill a tube, fill the balance of the volume with sterile PBS. Ultracentrifuge tubes can collapse if not filled almost to capacity and if not correctly balanced.
Weigh the buckets, writing down the weights so opposing buckets are balanced. Opposing buckets should be within 0.01 g of each other.
In the BSL-2 cabinet or clean bench, so the tubes are not contaminated, add PBS to the lighter tubes in buckets as necessary to balance the centrifuge rotor.
Transfer the rotor containing the samples into the ultracentrifuge, and centrifuge the supernatant at 100,000 × g and 4°C for 3 h.
For the centrifuging step, decant and discard the supernatant carefully and in sterile conditions. Use a micropipette with a sterile tip to remove the liquid that forms at the tip of the tube but does not drop off.
Note: Be careful not to contaminate the tube. In this step, free DiR goes away with the decanted supernatant, so the solution can be decanted, but always be careful not to lose the pellet.
Add 400 µL (360 µL) PI to each tube, and resuspend the pellet by gentle pipetting up and down. Add about 36.5 mL (35 mL) of PBS to each tube.
Weigh the buckets, and add PBS appropriately to balance the rotor to a precision of 0.01 g.
Centrifuge at 100,000 × g and 4°C for 18 h. Very carefully, decant the supernatant, and remove the residual liquid at the lip with a pipetter.
Resuspend the pellets with 100–150 µL PI (protease inhibitor cocktail) in PBS (see Recipes). Gently pipette the solution up and down. Use the tip to gently scrape around the bottom of the tube, to resuspend the vesicles. If desired, combine all samples into one microfuge tube.
Wash DiR-labeled exosomes in Amicon Ultra 4 with 10,000 MWCO, to remove free DiR. Add the sample to Amicon Ultra, and bring the volume to 4 mL total volume, using PBS supplemented with 1% (v/v) PI (i.e., if 2 mL is needed to bring to 4 mL total, add 2 mL of PBS and 20 µL of PI).
Centrifuge DiR-labeled exosomes in Amicon Ultra at 4,000 × g (in swinging bucket rotor, check × g limit if using a fixed angle rotor) at 4°C, in approximately 3-min intervals. Check the volume, and ensure the top chamber does not run dry because that is where the EVs are located. Reduce volume to about 250–400 µL. Discard the lower chamber filtrate and proceed to the next steps.
Transfer the sample containing DiR labeled exosomes in the upper chamber to a fresh microfuge tube, by gently pipetting and using the tip to gather as much liquid from the chamber as possible. Some free DiR may be seen as an insoluble precipitate.
To remove precipitated free DiR dye from DiR labeled exosomes, centrifuge the sample at 16,000 × g and 4°C for 20 min. Gently pipette the supernatant to a fresh microfuge tube.
Note: The supernatant may appear only very light blue to bluish at this stage.
Proceed to protein concentration determination using a micro BCA protein assay kit (Thermo Fisher). Use a nanoparticle characterization system (e.g., NanoSight LM10) to count the particles.
Performing EV uptake experiments
Prepare a 96-well plate of cells at 70% confluency the day before the cellular uptake experiment. The confluency required might depend on the cell type used.
Determine EV concentration through nanoparticle tracking analysis, such as NanoSight, to prepare a standardized number of EVs for cellular uptake experiments.
Prepare a 10-mL solution of 2e7 EVs (2e6/mL EVs) in serum-free OptiMEM containing appropriate antibiotics, for application to the cells.
Remove media from cells growing on a 96-well plate, and wash twice with sterile 1× PBS.
Add 100 µL of a prepared solution of 2e6/mL EVs in serum-free OptiMEM containing appropriate antibiotics to each well in a 96-well plate.
As a control for the DiR-labeled exosomes, a 500-µL solution of 5 µM DiR in OptiMEM can be used, in addition to the labeled exosome samples.
Cells can be imaged over 24 h using a Cytation5 instrument set to 5% CO2 and 37°C for every 2 h. See Figures 2 and 3, where L442 mouse-derived lymphocytes are used.
After imaging of the live cell uptake has been completed, the cells can be fixed with 4% PFA (paraformaldehyde) at room temperature for 15 min, and stained with DAPI in the dark at room temperature for 10 min to count DAPI objects, and normalize EV count to 1,000 cells.
Data and statistical analysis are performed using Microsoft Excel and GraphPad Prism.
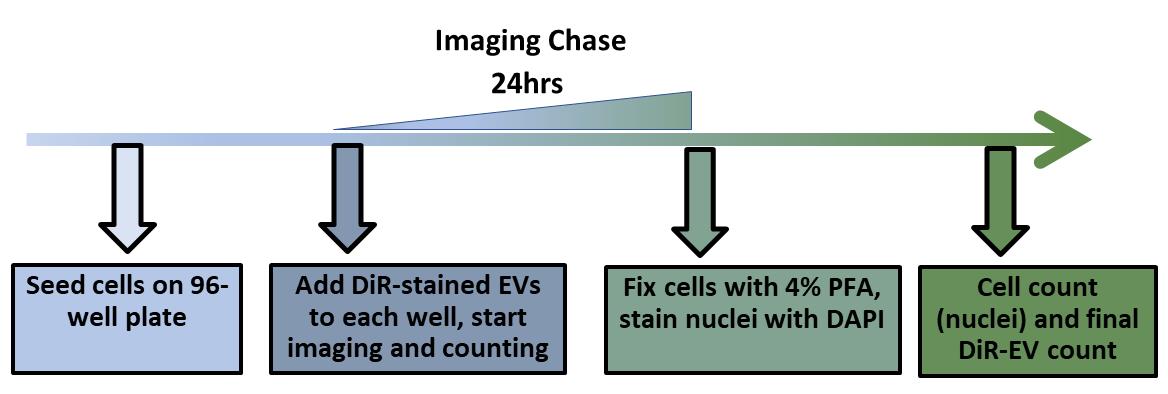
Figure 2. Experimental setup for the EV uptake experiment.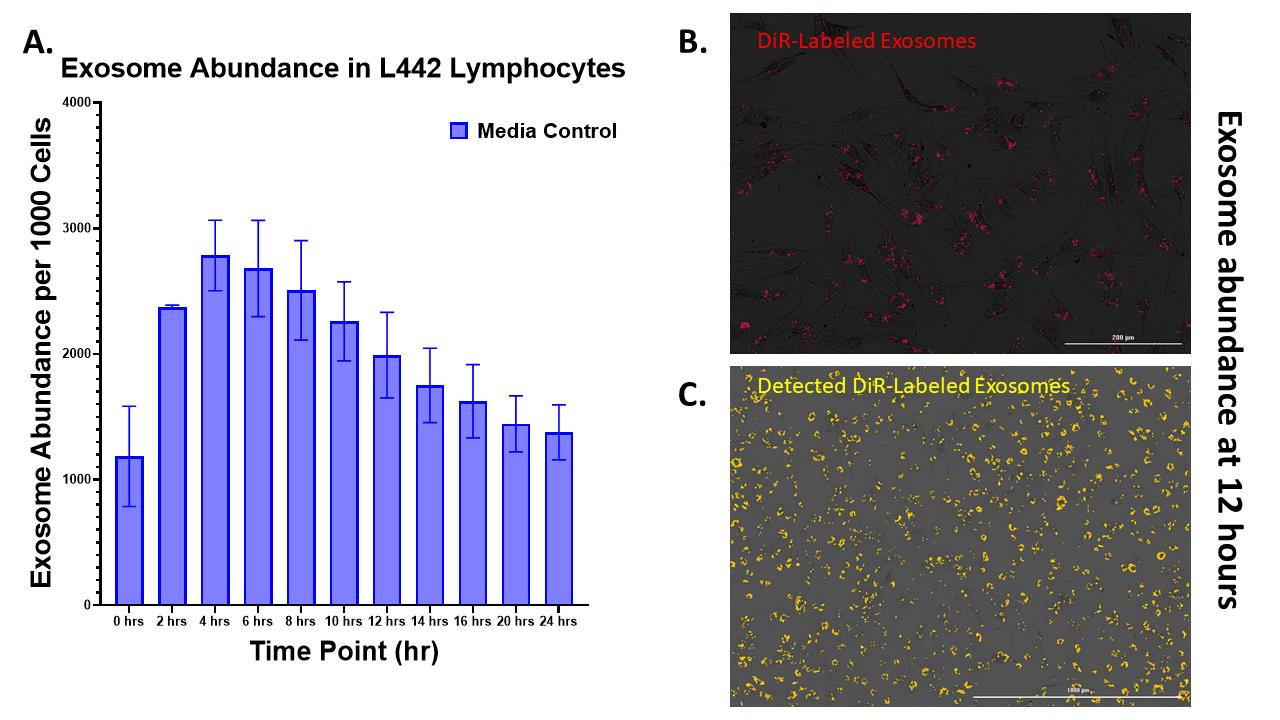
Figure 3. Quantifying the abundance of EVs in cells. (A) Labeled exosome abundance detected per 1,000 cells over 24-h, according to specific parameters in Cytation5. Three replicates of L442 mouse lymphocytes were prepared on a 96-well plate, and grown to 70% confluency prior to the addition of exosomes. A specific serum-free OptiMEM with antibiotics containing 2e6 labeled exosomes per mL was added to the cells. Cells were imaged under 10× magnification, with brightfield and the CY7 fluorescent filter, every 2 h up to the 16-h timepoint, then every 4 h up to 24 h after that. After the 24-h reading was complete, the cells were washed and fixed with 4% PFA, and stained with DAPI, to normalize the exosome count to 1,000 cells. The graph here shows the average exosome abundance across the three wells of L442 mouse lymphocytes (GraphPad Prism). (B) A representative image of fluorescent exosomes in L442 mouse lymphocytes at the 12-h time point. (C) A fully processed image of cellular uptake of exosomes with exosomes specifically detected in the Cytation5 software is highlighted by a yellow border. A 3 × 3 area of 10× magnified images was acquired, preprocessed, deconvoluted, and stitched for each well.
Recipes
Media containing exosome-depleted fetal bovine serum (FBS) and penicillin/streptomycin
In a BSL-2 cabinet, prepare 500 mL of OptiMEM with 3% (v/v) exosome-depleted FBS, and 1% (v/v) of penicillin/streptomycin antibiotic solution. The media can then be sterile filtered through a vacuum filter and stored at 4°C before use.
Control Sample for NanoSight quantification
Control can be prepared using DiR and PBS with 1% (v/v) PI.
From the stock DiR of 2.5 mg/mL, prepare a 5 µM dilution in the PBS and PI solution, then incubate in the dark at room temperature for 30 min, with occasional gentle inversion. Follow steps C11 through C13 of the procedure to clean and condense the control sample, as for the DiR-treated sample. After the sample has been prepared, it can be used in NanoSight experiments as a blank for the DiR-treated samples.
Protease inhibitor cocktail (PI) from Pierce Protease Inhibitor Tablets, EDTA-Free
A protease inhibitor cocktail (PI) can be prepared from Pierce Protease Inhibitor Tablets by dissolving one tablet per 5 mL of PBS to make 10% stock. This PI is added at 1% (v/v) to the conditioned media and to each wash step (for example, 500 µL of PI is added to 50 mL of conditioned media).
Acknowledgments
This work was supported by U. S. Public Health Grant R03 AI-135610 (MJE), R01 AI158749-02 (MJE), and 1R21NS113649-01 from the National Institutes of Health. The funders had no role in study design, data collection, analysis, decision to publish, or manuscript preparation.
Competing interests
The authors have declared that no competing interests exist.
References
- Busato, A., Bonafede, R., Bontempi, P., Scambi, I., Schiaffino, L., Benati, D., Malatesta, M., Sbarbati, A., Marzola, P. and Mariotti, R. (2016). Magnetic resonance imaging of ultrasmall superparamagnetic iron oxide-labeled exosomes from stem cells: a new method to obtain labeled exosomes.Int J Nanomedicine 11: 2481-2490.
- Busato, A., Bonafede, R., Bontempi, P., Scambi, I., Schiaffino, L., Benati, D., Malatesta, M., Sbarbati, A., Marzola, P. and Mariotti, R. (2017). Labeling and Magnetic Resonance Imaging of Exosomes Isolated from Adipose Stem Cells. Curr Protoc Cell Biol 75: 3.44.41-43.44.15.
- Cook, R. L., Householder, K. T., Chung, E. P., Prakapenka, A. V., DiPerna, D. M. and Sirianni, R. W. (2015). A critical evaluation of drug delivery from ligand modified nanoparticles: Confounding small molecule distribution and efficacy in the central nervous system. J Control Release 220(Pt A): 89-97.
- Corso, G., Heusermann, W., Trojer, D., Görgens, A., Steib, E., Voshol, J., Graff, A., Genoud, C., Lee, Y., Hean, J., et al. (2019). Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule - single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J Extracell Vesicles 8(1): 1663043.
- Gangadaran, P., Li, X. J., Lee, H. W., Oh, J. M., Kalimuthu, S., Rajendran, R. L., Son, S. H., Baek, S. H., Singh, T. D., Zhu, L., et al. (2017). A new bioluminescent reporter system to study the biodistribution of systematically injected tumor-derived bioluminescent extracellular vesicles in mice. Oncotarget 8(66): 109894-109914.
- Huotari, J. and Helenius, A. (2011). Endosome maturation. EMBO J 30(17): 3481-3500.
- Somanchi, S. S. (2016). Noninvasive In Vivo Fluorescence Imaging of NK Cells in Preclinical Models of Adoptive Immunotherapy. Methods Mol Biol 1441: 307-316.
- Théry, C., Zitvogel, L. and Amigorena, S. (2002). Exosomes: composition, biogenesis and function. Nat Rev Immunol 2(8): 569-579.
- Zhao, Q., Hai, B., Kelly, J., Wu, S. and Liu, F. (2021). Extracellular vesicle mimics made from iPS cell-derived mesenchymal stem cells improve the treatment of metastatic prostate cancer. Stem Cell Res Ther 12(1): 29.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Santelices, J., Ou, M., Hui, W. W., Maegawa, G. H. B. and Edelmann, M. J. (2022). Fluorescent Labeling of Small Extracellular Vesicles (EVs) Isolated from Conditioned Media. Bio-protocol 12(12): e4447. DOI: 10.21769/BioProtoc.4447.
Category
Cell Biology > Organelle isolation > Extracellular vesicle
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link