- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Time-off-pick Assay to Measure Caenorhabditis elegans Motility
Published: Vol 12, Iss 12, Jun 20, 2022 DOI: 10.21769/BioProtoc.4436 Views: 3176
Reviewed by: Juan Facundo Rodriguez AyalaMatthias RieckherRajesh RanjanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Plate Assay to Determine Caenorhabditis elegans Response to Water Soluble and Volatile Chemicals
Takashi Murayama and Ichiro N. Maruyama
Feb 20, 2018 9047 Views

Measuring Spatiotemporal Dynamics of Odor Gradient for Small Animals by Gas Chromatography
Akiko Yamazoe-Umemoto [...] Koutarou D. Kimura
Apr 5, 2018 7414 Views

Aerotaxis Assay in Caenorhabditis elegans to Study Behavioral Plasticity
Qiaochu Li [...] Karl Emanuel Busch
Aug 20, 2022 2251 Views
Abstract
Caenorhabditis elegans is a simple metazoan that is often used as a model organism to study various human ailments with impaired motility phenotypes, including protein conformational diseases. Numerous motility assays that measure neuro-muscular function have been employed using C. elegans. Here, we describe “time-off-pick" (TOP), a novel assay for assessing motility in C. elegans. TOP is conducted by sliding an eyebrow hair under the mid-section of the worm and counting the number of seconds it takes for the worm to crawl completely off. The time it takes for the worm to crawl off the eyebrow hair is proportional to the severity of its motility defect. Other readouts of motility include crawling or swimming phenotypes, and although widely established, have some limitations. For example, worms that are roller mutants are less suitable for crawling or swimming assays. We demonstrated that our novel TOP assay is sensitive to age-dependent changes in motility, thus, providing another more inclusive method to assess motor function in C. elegans.
Graphical abstract:

Conceptual overview of the “time-off-pick” (TOP) assay. Various C. elegans models exhibit age-dependent defects in motility. The time it takes for a worm to crawl off of an eyebrow pick that is slid under its mid-section is measured in TOP seconds. A greater TOP is indicative of a greater motility defect. Eventually, worms with phenotypes that lead to paralysis will not be able to leave the pick.
Background
Caenorhabditis elegans is a 1-mm long nematode often used as a model organism for studying various human diseases or conditions that induce neuro-muscular defects. As such, motility assays are commonly used to assess motor impairments in C. elegans. Common motility assays measure parameters associated with swimming and crawling phenotypes, which have proven to be very successful and effective readouts (Cohen et al., 2012; Winter et al., 2016). However, the aforementioned assays can be time-consuming, may require specialized equipment, and are less compatible with certain C. elegans phenotypes, such as the roller phenotype, which is commonly used as a selection marker when generating transgenic C. elegans lines. The protocol presented in this paper, “time-off-pick” (TOP), describes a novel phenotypic assay for measuring motility in C. elegans. Our method is less elaborate, does not require specialized equipment, and is more inclusive to a variety of genetic backgrounds, including those that exhibit the roller phenotype. TOP involves sliding an eyebrow hair under the mid-section of the worm and counting the number of seconds it takes for the worm to crawl off (Video 1). The worm is not lifted by the eyebrow hair; rather, the hair remains stationary after being gently placed between the worm and the surface of the agar on nematode-growth media (NGM) plates, with care taken not to poke the side of the worm with the eyebrow hair. A longer TOP is indicative of a greater motility impairment. This method allows the experimenter to study C. elegans strains, mutations, conditions, or treatments that alter motility.
To demonstrate the feasibility of our method, we used C. elegans strains that are known to affect motility: lines carrying transgenic polyglutamine (polyQ) tracts (Morley et al., 2002; Brignull et al., 2006; Mohri-Shiomi and Garsin, 2008) and a mutant strain that manifests in a temperature-sensitive motility defect (Macleod et al., 1977). PolyQ tracts aggregate in a polyQ length- and an age-dependent manner, and increased polyQ aggregation is associated with increased motility defects (Morley et al., 2002; Brignull et al., 2006; Mohri-Shiomi and Garsin, 2008). In our previous work, we demonstrated that the results obtained from the TOP assay matched those from the swimming assay (“thrashing”) (Walker et al., 2021). Here, we show our TOP method can successfully assess changes in age- and polyQ length-dependent motility in worms harboring polyQ tracts in the intestine (Figure 1A), muscle (Figure 1B), and neurons (Figure 1C). To demonstrate the versatility of our method, we used a strain (CB1301) that expresses a temperature-sensitive mutation in myosin heavy chain gene, unc-54 (e1301), that results in impaired motility at restrictive temperatures (Macleod et al., 1977; Gidalevitz et al., 2006). We found that TOP detected a temperature-induced motility impairment 1 h after worms were placed at the restrictive temperature (Figure 1D). Together, these results demonstrate the use and efficacy of the TOP phenotype, which provides a validated and sensitive addition to existing motility assays.
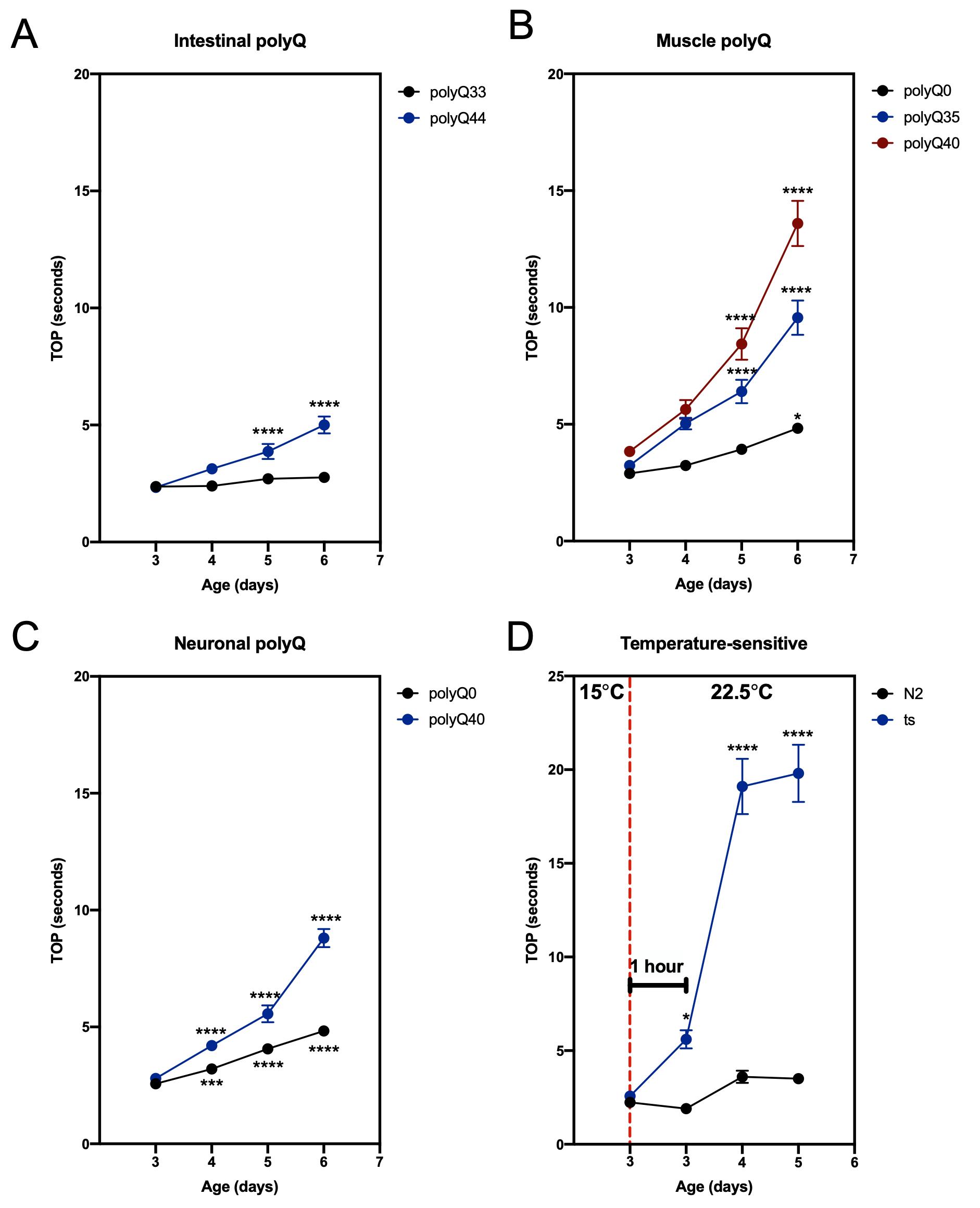
Figure 1. TOP measurement in C. elegans expressing polyQ and a temperature-sensitive mutation. (A–C) The TOP phenotype positively correlates with C. elegans age and polyQ length in the (A) intestine (rollers): polyQ44: AM738 rmIs297[vha-6p::q44::yfp; rol-6 (su1006)]; polyQ33: AM712 rmIs281[vha-6p::q33::yfp; rol-6 (su1006)], (B) muscle: polyQ40: AM141 rmIs133[unc-54p::q40::yfp]; polyQ35: AM140 rmIs132[unc-54p::q35::yfp]; polyQ0: AM134 rmIs126[unc-54p::q0::yfp], and (C) neurons: polyQ40: AM101 rmIs110[F25B3.3P::q40::yfp]; polyQ0: AM52 rmIs182[F25B3.3p::q0::yfp]. Worms were cultured and maintained at 22.5°C. (D) Motility of worms (CB1301) expressing a temperature-sensitive mutation (ts) in myosin heavy chain, unc-54(e1301). Nematodes cultured and maintained at 20°C harboring the mutation display a significant increase in TOP (seconds) after 1 h at a restrictive temperature of 22.5°C (data point immediately to the right of the red, dashed line), whereas wild-type worms (N2), exhibit no significant change. Motility impairment continues to worsen over the next 48 h at the restrictive temperature. Data are represented as the average TOP per worm. Each data point is an average of two independent experiments with a total of 30 worms. Error bars represent standard error of the mean (SEM). Statistical analysis was calculated using one-way analysis of variants (ANOVA) followed by multiple comparison Dunnett’s post-hoc test (*P < 0.05, ***P < 0.001, **** P < 0.0001).
Materials and Reagents
Note: Unless otherwise specified, all reagents are stored at room temperature.
Erlenmeyer flasks (2,000 mL and 1,000 mL)
6 cm Petri dishes (Genesee Scientific, catalog number: 32-105G)
Magnetic stir bar
Autoclave tape
Caenorhabditis elegans
500 mL Olympus vacuum filter flasks (Genesee Scientific, catalog number: 25-227)
Agar (Fisher Scientific, catalog number: BP1423)
Double distilled water (ddH2O)
NaCl (Fisher Scientific, catalog number: S671-500)
Trypticase-peptone (Gibco, catalog number: 211921)
Cholesterol (MP Biomedicals, catalog number: ICN10138201)
Ethanol, 200-proof (Decon Laboratories, catalog number: 04-355-223)
CaCl2·2H2O (Fisher Scientific, catalog number: C79-500)
MgSO4·7H2O (Fisher Scientific, catalog number: A14491)
KH2PO4 (Fisher Chemical, catalog number: P285-3)
NA2HPO4·7H2O (Fisher Scientific, catalog number: S373-500)
M9 Minimal Medium (see Recipes)
Cholesterol Stock Solution (see Recipes)
1 M CaCl2 Stock Solution (see Recipes)
1 M MgSO4 Stock Solution (see Recipes)
1 M KH2PO4, pH 6.0 Stock Solution (see Recipes)
Eyebrow hair pick (see Recipes)
NGM Plates (see Recipes)
Equipment
Zeiss Stemi 305 stereo microscope (Zeiss, catalog number: 435063-9010-000)
Incubators for C. elegans (15–25°C)
Ethanol candle (DWK Life Sciences, catalog number: 04-245-1)
Autoclave
pH meter (Thermo Electric Corporation)
Fisher Scientific Isotemp Stirrer (Fisher Scientific, catalog number: 11-100-49S)
Pasteur pipette (Fisher Scientific, catalog numer: 22-378893)
Soft and thin eyebrow hair
Tape
Second counter (such as a YouTube video or app that beeps every second)
Platinum wire worm pick (made in-house as described in Wollenberg et al., 2013)
Software
GraphPad Prism v8.4.3. (GraphPad Software, Inc.)
BioRender (www.biorender.com)
Procedure
Notes:
Unless otherwise specified, all steps are conducted outside of a biological safety cabinet (BSC) using aseptic techniques.
This protocol assumes nematodes have already been cultured and grown to the age of interest on solid nematode growth media (NGM) plates.
Nematodes used for the data presented herein were cultured and maintained on E. coli OP50 and in accordance with methods we have used previously (Walker et al., 2021) .
C. elegans size will affect TOP measurements; therefore, we recommend using this assay on adult worms only. Also, the experimenter has to use proper size-matched controls when testing conditions that affect worm size.
Worm preparation
To reduce error, transfer worms using platinum wire pick onto a new NGM plate seeded with bacteria of interest. Make sure there is an equal number of worms per plate as well as an equal amount of bacteria on each plate that is consistent between test and control worms.
Note: Make sure to flame the worm pick between samples to avoid cross-contaminating with bacteria or worm strains.
Motility assessment
Note: This step is performed using a dissection microscope and a ticking second timer.
Using a pick with an eyebrow hair taped to it (Figure 2), put an NGM plate that contains worms in view. Slide the eyebrow hair under the mid-section of a worm and count the number of seconds it takes for the worm to crawl off entirely.
Note: It is important to make sure that the worm is not poked in the side with the eyebrow hair; instead, the hair should delicately be placed gently underneath the worm. Additionally, ensure that the eyebrow hair is not going into the agar, is on top of the agar at all times, and remains stationary between the surface of the agar and the worm. Do not lift the worm with the eyebrow hair.
Record the number of seconds it takes the worm to crawl off. Continue with worms and samples as desired.
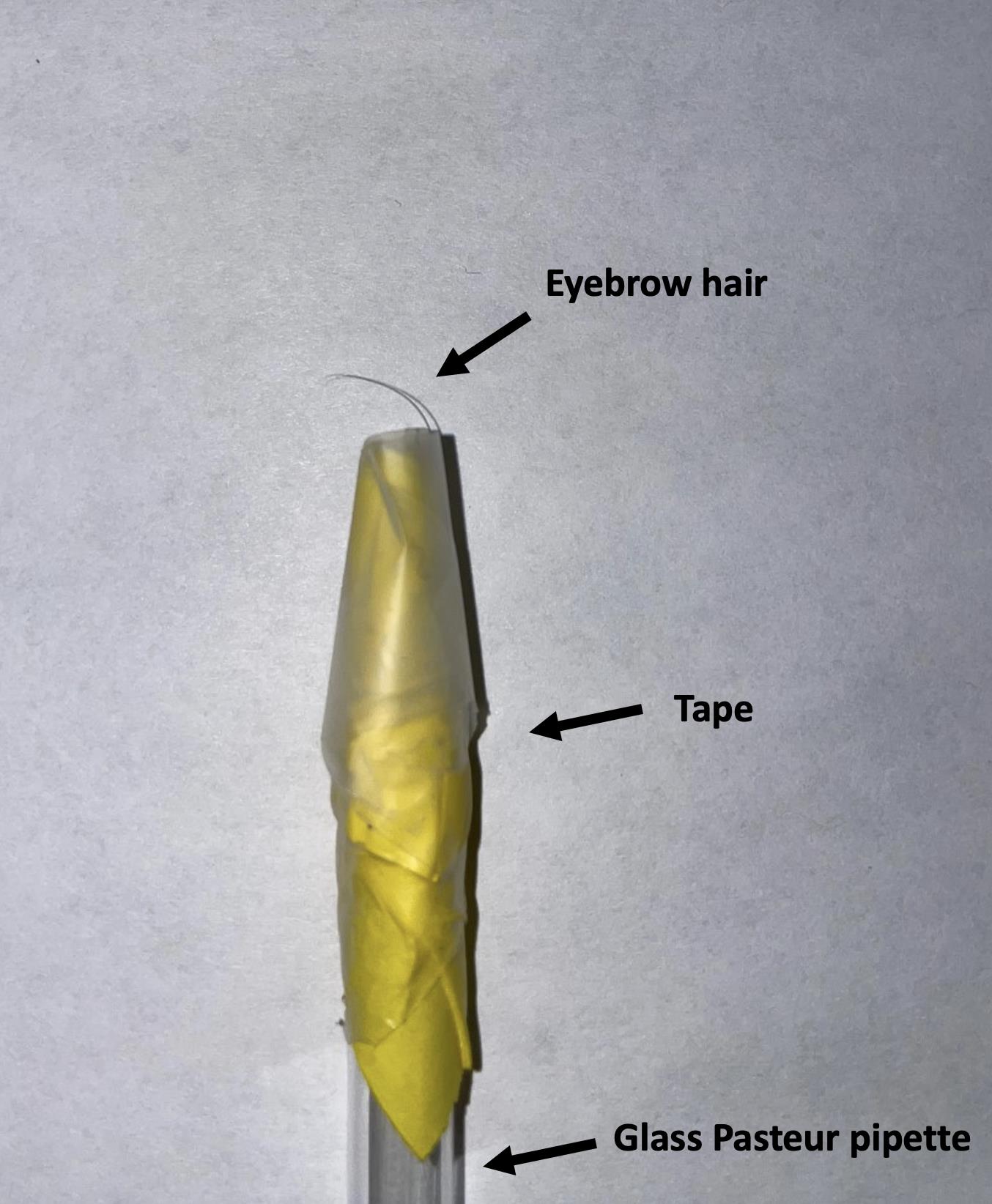
Figure 2. Picture of the eyebrow hair-pick used in the TOP assay.
Data analysis
The TOP assay allows the experimenter to assess changes in motility by measuring the number of seconds it takes for a worm to completely move off an eyebrow pick. The TOP measurement is proportional to motility—the more time it takes, the larger the defect. The results are analyzed by comparing the measurements of the experimental group with those of the control. We used various C. elegans strains to demonstrate the feasibility of the TOP assay. Data are representative of two independent experiments with a minimum of 30 worms total. The data are represented as standard error of the mean (SEM), and the statistical significance was calculated using student’s t-test (see legend in Figure 1).
Notes
All reagents should be sterile to avoid contamination of samples. All other precautionary notes are contained within corresponding steps of the protocol in “Procedure”.
Recipes
M9 Minimal Medium
Combine 5.8 g of Na2HPO4·7H2O, 3.0 g of KH2PO4, 5.0 g of NaCl, and 0.25 g of MgSO4·7H2O in a 1 L flask.
Add a magnetic stir bar and fill to 1 L with ddH2O.
Place on stir plate and let mix until all reagents are dissolved.
Filter sterilize into two 500 mL bottles.
Aliquot into smaller sterile bottles if desired, and store at RT.
Cholesterol Stock Solution
Dissolve cholesterol in 200-proof ethanol to a final concentration of 5 mg/mL cholesterol. Store at -20°C until use.
1 M CaCl2 Stock Solution
Add 147 g of CaCl2·2H2O to a 1 L flask.
Fill to 1 L with ddH2O.
Add a magnetic stir bar.
Place on stir plate until reagent dissolves.
Aliquot and screw caps loosely on bottles.
Adhere autoclave tape.
Autoclave fluid cycle for 30 min.
Let cool, fully screw caps on, and store at RT until use.
1 M MgSO4 Stock Solution
Add 246.5 g of MgSO4·7H2O to a 1 L flask.
Fill to 1 L with ddH2O.
Add a magnetic stir bar.
Place on stir plate until reagent dissolves.
Aliquot and screw caps loosely on bottles.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Let cool, fully screw caps on, and store at RT until use.
1 M KH2PO4, pH 6.0 Stock Solution
Add 136.1 g of KH2PO4 to a 1 L flask.
Dissolve in 800 mL of ddH2O.
Add a magnetic stir bar.
Place on stir plate until reagent dissolves.
Adjust pH to 6.0 with 10 g of KOH. Add ddH2O up to 1 L.
After reagents are completely dissolved, and pH is adjusted to 6.0, aliquot and screw caps loosely on bottles.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Let cool, fully screw caps on, and store at RT until use.
Eyebrow hair pick
Place soft and relatively thin eyebrow hair in the smaller tip of a glass Pasteur pipette, root-first. Secure with tape.
NGM Plates
In a 2 L flask, combine 3.0 g of NaCl, 2.5 g of trypticase-peptone, and 17.0 g of agar.
Fill ddH2O up to 1 L.
Add a magnetic stir bar.
Place on stir plate and let reagents mix for several minutes.
Place cap (loosely screwed on) or foil on the opening of the bottle.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Cool to 50°C.
Add 1 mL of 5 mg/ mL cholesterol stock solution, 1 mL of 1 M CaCl2 stock solution, 1 mL of 1 M MgSO4 stock solution, and 25 mL of 1M KH2PO4, pH 6.0 stock solution.
Note: Let cholesterol stock solution (5 mg/mL) reach room temperature and ensure cholesterol re-dissolves in solution before adding to NGM.
Cover and put back on stir plate for one minute.
Pour into 60 mm Petri dishes.
Let cool and store upside down in a plastic box, with lid at 4°C.
Acknowledgments
This work was supported in part by the Infectious Diseases Society of America, The National Institutes of Health (R03AG070580-01), and Start-up funding provided by the Microbiology and Cell Science Department at the University of Florida Institute of Food and Agricultural Sciences to DMC. Walker et al. (2021) is the original research paper from where this protocol was derived. The Graphical Abstract figure was created using BioRender.com.
Competing interests
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interests exist.
References
- Cohen, E., Yemini, E., Schafer, W., Feitelson, D. G. and Treinin, M. (2012). Locomotion analysis identifies roles of mechanosensory neurons in governing locomotion dynamics of C. elegans. J Exp Biol 215(Pt 20): 3639-3648.
- Winter, P. B., Brielmann, R. M., Timkovich, N. P., Navarro, H. T., Teixeira-Castro, A., Morimoto, R. I. and Amaral, L. A. (2016). A network approach to discerning the identities of C. elegans in a free moving population. Sci Rep 6: 34859.
- Mohri-Shiomi, A. and Garsin, D. A. (2008). Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem 283(1): 194-201.
- Morley, J. F., Brignull, H. R., Weyers, J. J. and Morimoto, R. I. (2002). The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A 99(16): 10417-10422.
- Brignull, H. R., Moore, F. E., Tang, S. J. and Morimoto, R. I. (2006). Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J Neurosci 26(29): 7597-7606.
- MacLeod, A. R., Waterston, R. H., Fishpool, R. M. and Brenner, S. (1977). Identification of the structural gene for a myosin heavy-chain in Caenorhabditis elegans. J Mol Biol 114(1): 133-140.
- Walker, A. C., Bhargava, R., Vaziriyan-Sani, A. S., Pourciau, C., Donahue, E. T., Dove, A. S., Gebhardt, M. J., Ellward, G. L., Romeo, T. and Czyz, D. M. (2021). Colonization of the Caenorhabditis elegans gut with human enteric bacterial pathogens leads to proteostasis disruption that is rescued by butyrate. PLoS Pathog 17(5): e1009510.
- Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R. and Morimoto, R. I. (2006). Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311(5766): 1471-1474.
- Wollenberg, A. C., Visvikis, O., Alves, A.-M. F. and Irazoqui, J. E. (2013). Staphylococcus aureus Killing Assay of Caenorhabditis elegans. Bio-protocol 3(19): e916.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Walker, A. C., Bhargava, R., Brust, A. S., Owji, A. A. and Czyz, D. M. (2022). Time-off-pick Assay to Measure Caenorhabditis elegans Motility. Bio-protocol 12(12): e4436. DOI: 10.21769/BioProtoc.4436.
Category
Neuroscience > Behavioral neuroscience > Chemotaxis
Biological Sciences > Biological techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link