- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Growth Curve Measurements with a Multimode Microplate Reader
Published: Vol 12, Iss 9, May 5, 2022 DOI: 10.21769/BioProtoc.4410 Views: 7735
Reviewed by: Alba BlesaChangyi ZhangElías Barquero-CalvoDamián Lobato-Márquez

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

SIMBA Method—Simultaneous Detection of Antimicrobial and Anti-biofilm Activity of New Compounds Using Salmonella Infantis
Meta Sterniša [...] Anja Klančnik
Aug 5, 2023 2008 Views

Functional Assay for Measuring Bacterial Degradation of Gemcitabine Chemotherapy
Serkan Sayin and Amir Mitchell
Sep 5, 2023 1741 Views

Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology
Chen Tang [...] Guangxin Xiang
Feb 5, 2025 1985 Views
Abstract
Bacterial studies based on growth curves are common in microbiology and related fields. Compared to the standard photometer and cuvette based protocols, bacterial growth curve measurements with microplate readers provide better temporal resolution, higher efficiency, and are less laborious, while analysis and interpretation of the microplate-based measurements are less straightforward. Recently, we developed a new analysis method for evaluating bacterial growth with microplate readers based on time derivatives. Here, we describe a detailed protocol for this development and provide the homemade program for the new analysis method.
Keywords: Time derivativeBackground
Monitoring the growth and proliferation of microorganisms based on growth curves is commonly used in microbiology and related fields for understanding the growth of microorganisms, the efficacy of antibiotic drugs, and the microbial responses to new environmental conditions and stresses (Mandelstam et al., 1982; Andrews, 2001; Bollenbach et al., 2009; Haque et al., 2017). Traditional growth curve measurements based on photometers and cuvettes suffer from several drawbacks, such as low temporal resolution, low efficiency, and high labor requirement (Stevenson et al., 2016; Goldman and Green, 2021). In contrast, growth curve measurements using microplate readers take advantage of automation and parallel capability; however, the application of microplate readers for growth curve measurements has been hindered due to the complexity in the quantification and data interpretation caused by multiple light scattering and/or light absorption/scattering by other ingredients in the samples (Stevenson et al., 2016). Recently, we developed a new analysis method for evaluating bacterial growth with microplate readers based on time derivatives. This method not only uses the optical density (i.e., absorption/scattering) of bacterial culture, but also takes advantage of the fluorescence of bacteria expressing fluorescent proteins (Krishnamurthi et al., 2021). The time derivatives of optical density and fluorescence show peaks, whose location and height are correlated with the lag time and maximum growth rate of bacteria (Krishnamurthi et al., 2021). Here, we describe the detailed protocol for carrying out growth curve measurements with microplate readers and provide the homemade program for analyzing and visualizing the results. Escherichia coli bacteria were used here as an example; however, this protocol is expected to be applicable to other microorganisms.
Materials and Reagents
96-well clear bottom plates (Corning, catalog number: 3631) (Figure 1)

Figure 1. Photo of a 96-well microplate with clear bottom and its clear lid.Polystyrene lids for 96-well microplates (Corning, catalog number: 3931)
1–200 µL pipette tips (VWR, catalog number: 53508-810)
100–1,000 µL pipette tips (VWR, catalog number: 83007-376)
1.5 mL microcentrifuge tubes (VWR, catalog number: 20170-038)
14 mL sterile culture tubes (VWR, catalog number: 60818-725)
Escherichia coli strain K-12 MG1655 transformed with plasmid pOEGFP2, encoding ampicillin resistance gene and green fluorescent proteins (Lab collection), which were used previously (Haque et al., 2017; Niyonshuti et al., 2020; Krishnamurthi et al., 2021). (Figure 2)
Note: Other bacterial strains expressing fluorescent proteins are expected to be compatible with the current protocol.
Figure 2. Map of the plasmid pOEGFP2 used in this protocol. The plasmid was a gift from Dr. David McMillen at the University of Toronto. The plasmid map was generated using SnapGene Viewer (GSL Biotech LLC).Tryptone (Bio Basic Inc, catalog number: TG217)
Yeast extract (Bio Basic Inc, catalog number: G0961)
Sodium chloride (VWR International, LLC, catalog number: 7647-14-5)
Ampicillin (VWR, catalog number: AAJ60977-06)
Triton X-100 (VWR, catalog number: AAA16046-AP)
Ethanol (VWR, catalog number: 89125-188)
Deionized water (>17.6 MΩ, from Barnstead ultrapure water purification system [model: D4641])
Triton X-100 (VWR, catalog number: AAA16046-AP)
Liquid LB medium (200 mL) (see Recipes)
Ampicillin stock solution (10 mL) (see Recipes)
Python code (see Supplementary file)
Equipment
2–20 µL single channel mechanical pipette (VWR, catalog number: 89079-964)
20–200 µL single channel mechanical pipette (VWR, catalog number: 89079-970)
Multimode microplate reader capable of measuring both absorption and fluorescence (BioTek Instruments, model: Synergy H1)
Autoclave (Tuttnauer, model: 2540E-B/L)
Biosafety cabinet (Labconco, model: Purifier Logic+ Class II Type A2)
Software
Gen5 Microplate Reader and Imager Software (BioTek Instruments, https://www.biotek.com/)
Microsoft Excel Spreadsheet Software (Microsoft Corporation, https://www.microsoft.com/)
Python (https://www.python.org/)
Procedure
Bacteria Growth:
Preparation of initial liquid culture of bacteria
Prepare liquid LB medium, followed by autoclaving (120°C, 41 min) and cooling down to room temperature.
Inoculate a single colony of the bacterial strain into 7 mL of LB medium in a culture tube, supplemented with ampicillin or the appropriate antibiotic at a final concentration of 50 µg/mL.
Note: Several colonies in separate culture tubes may be used if more replicates are desired.Incubate the liquid culture in a shaking incubator at 37°C with orbital shaking at 250 rpm overnight.
Pre-treatment of 96-well plates and lids
Prepare 10% Triton X-100 in ethanol by mixing 1 mL of Triton X-100 with 9 mL of ethanol.
Pour 10 mL of the diluted Triton X-100 to a new microplate lid.
Incubate the lid with diluted Triton X-100 at room temperature for 15 s.
Take a new 96-well plate and fill each well with 100% ethanol (400 µL per well). Incubate at room temperature for 15 min.
Discard ethanol from the microplate wells and discard the diluted Triton X-100 from the microplate lid.
Air dry the microplate wells and microplate lid at room temperature by leaving them inside the biosafety cabinet for 30 min.
Turn on the UV light of the biosafety cabinet and further disinfect the microplate wells and lid for 15 min.
Note: Treatment with ethanol and UV is for sterilization, while treatment with Triton X-100 is to prevent water condensation on the lid during the measurements.
Setup of microplate reader and software
Turn on the microplate reader and open the Gen5 software.
Set up a new procedure/protocol in Gen5 (Figure 3).
Plate type: 96-well plate.
Use lid: checked.
Temperature: 37°C.
Shake: orbital for 0:10.
Kinetic measurement for a desired total time (e.g., 16 h, 24 h, or 48 h) and interval of 10 min with continuous orbital shaking.
Reading: absorption at 600 nm, fluorescence at excitation = 488 nm and emission = 525 nm.
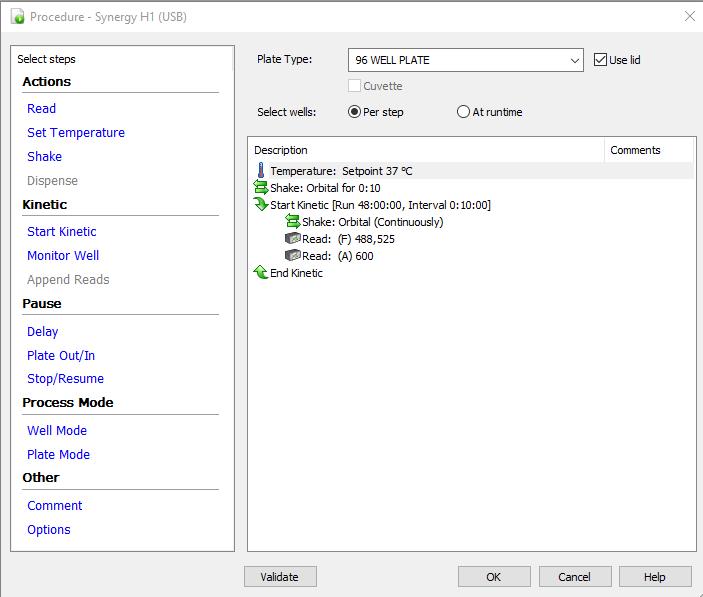
Figure 3. An example for setting up the procedure/protocol in Gen5 software for the growth curve measurements of bacteria using a microplate reader.
Growth curve measurements using microplate reader
Dilute the overnight liquid bacterial culture by 100–500 times in fresh LB supplemented with ampicillin at 50 µg/mL.
Notes:
If a photometer is available, measure the optical density of the overnight culture and make the dilution to the desired optical density (typically <0.05).
Other media or additional ingredients (such as ions, nanoparticles, and drugs of interest that may affect bacterial growth) can be used if desired. For example, silver ions were added to the LB medium at a final concentration of 40 µg/mL in the sample S2 of the representative data (Figure 4).
Pipette 200 μL of the diluted culture into a single well of the 96-well plate as a sample. Samples should be prepared with at least triplicates.
Pipette 200 µL of LB medium with ampicillin into a single well of the 96-well plate as a blank. Blanks should be prepared with at least triplicates.
After loading all the samples and blanks, cover the 96-well microplate with the prepared lid.
Start the procedure/protocol in Gen5 software. When prompted, place the microplate properly on the microplate holder, and start recording of the bacterial growth curve.
Finishing up
Upon completion of the data acquisition, remove the 96-well plate from the microplate reader.
Disinfect the samples and discard the microplate properly.
Save the results in Gen5 software, and export the data as an Excel spreadsheet file.
Close the Gen5 software, and turn off the microplate reader.
Data analysis
Reorganize the data from the Excel spreadsheet as text files so that each text file contains the absorption data (or fluorescence) data of a single sample (or the blank). Note that each column in the text file corresponds to the readings of a single well of the microplate. Fluorescence data and absorption data should be separated into different files.
Run the provided Python code (see Supplementary file) to:
Calculate the averaged absorption (or fluorescence) of samples, and plot the results.
Smooth the growth curves, calculate the averaged first-order time derivatives of absorption (or the second-order time derivative of fluorescence) of samples, and plot the results.
Identify the peak locations and heights of the time derivatives.
Compare the peak locations (and/or heights) to quantify the relative changes between the samples.
Representative Data
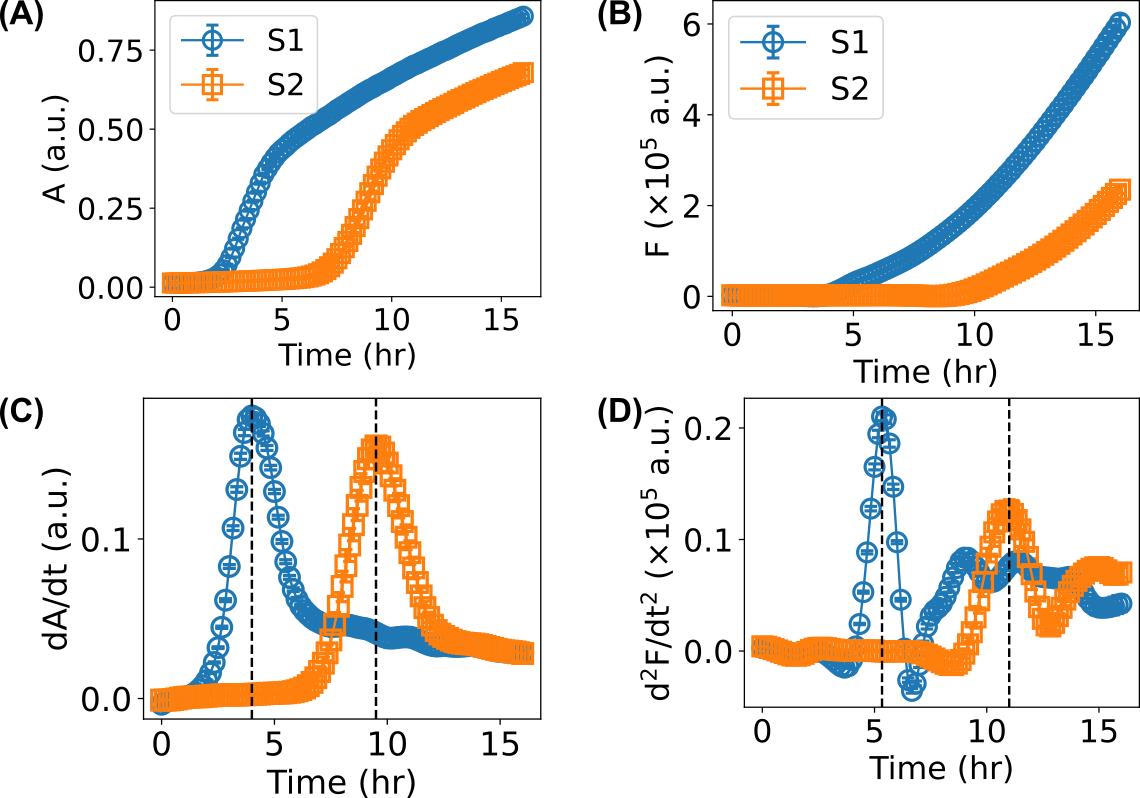
Figure 4. Representative data for the growth curves of E. coli bacteria measured by a microplate reader and their time derivatives. A. Absorption growth curves. B. Fluorescence growth curves. C. First-order time derivative of absorption growth curves. D. Second-order time derivative of fluorescence growth curves. Sample S1: MG1655 strain with pEOGFP2 plasmid in LB medium. Sample S2: MG1655 strain with pEOGFP2 plasmid in LB medium with silver ions of 40 µg/mL. In all sub-figures, “a.u.” stands for arbitrary units.
Recipes
Liquid LB medium (200 mL)
2 g Tryptone
1 g yeast extract
2 g sodium chloride (NaCl)
Fill up to 200 mL with deionized water and autoclave to sterilize.
Ampicillin stock solution (50 mg/mL)
500 mg of ampicillin powder (sodium salt)
10 mL of water
Filter sterilize the ampicillin solution and store 1 mL tubes at -20°C.
Acknowledgments
We thank Dr. David McMillen at the University of Toronto for the generous gift of plasmid encoding the enhanced GFP and ampicillin resistance (pOEGFP2). This work was supported by the University of Arkansas, the Arkansas Biosciences Institute (Grant No. ABI-0189, No. ABI-0226, No. ABI-0277, No. ABI-0326, No. ABI-2021, No. ABI-2022), and the National Science Foundation (Grant No. 1826642). The original protocol was developed and used in Krishnamurthi et al. (2021) and Niyonshuti et al. (2020).
Competing interests
No competing interest is declared.
References
- Andrews, J. M. (2001). Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48 Suppl 1: 5-16.
- Bollenbach, T., Quan, S., Chait, R. and Kishony, R. (2009). Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139(4): 707-718.
- Goldman, E., and Green, L. H. (2021). Practical Handbook of Microbiology (Taylor & Francis Group). CRC Press. IBSN: 9780367567637.
- Haque, M. A., Imamura, R., Brown, G. A., Krishnamurthi, V. R., Niyonshuti, I. I., Marcelle, T., Mathurin, L. E., Chen, J. and Wang, Y. (2017). An experiment-based model quantifying antimicrobial activity of silver nanoparticles on Escherichia coli. RSC Advances 7(89): 56173-56182.
- Krishnamurthi, V. R., Niyonshuti, II, Chen, J. and Wang, Y. (2021). A new analysis method for evaluating bacterial growth with microplate readers. PLoS One 16(1): e0245205.
- Mandelstam, J., Dawes, I. W., and McQuillen, K. (1982). Biochemistry of bacterial growth. New York, Wiley.
- Niyonshuti, II, Krishnamurthi, V. R., Okyere, D., Song, L., Benamara, M., Tong, X., Wang, Y. and Chen, J. (2020). Polydopamine Surface Coating Synergizes the Antimicrobial Activity of Silver Nanoparticles. ACS Appl Mater Interfaces 12(36): 40067-40077.
- Stevenson, K., McVey, A. F., Clark, I. B. N., Swain, P. S. and Pilizota, T. (2016). General calibration of microbial growth in microplate readers. Sci Rep 6: 38828.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rogers, A. T., Bullard, K. R., Dod, A. C. and Wang, Y. (2022). Bacterial Growth Curve Measurements with a Multimode Microplate Reader. Bio-protocol 12(9): e4410. DOI: 10.21769/BioProtoc.4410.
Category
Microbiology > Antimicrobial assay > Antibacterial assay
Biological Sciences > Microbiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









