- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Apoplastic Expression of CARD1-ecto Domain in Nicotiana benthamiana and Purification from the Apoplastic Fluids
Published: Vol 12, Iss 8, Apr 20, 2022 DOI: 10.21769/BioProtoc.4387 Views: 3557
Reviewed by: Ashish RanjanMalgorzata LichockaDemosthenis ChronisShweta Panchal

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2855 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1975 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1763 Views
Abstract
The protein expression and purification process is an essential initial step for biochemical analysis of a protein of interest. Traditionally, heterologous protein expression systems (such as E. coli, yeast, insect cells, and cell-free) are employed for plant protein expression, although a plant expression system is often desirable for plant proteins, to ensure proper post-translational modifications. Here, we describe a method to express and purify the ectodomain of one of the leucine-rich repeat receptor-like kinase called CARD1/HPCA1, from Nicotiana benthamiana apoplastic fluid. First, we express His-tagged CARD1 ectodomain in the apoplastic space of N. benthamiana by the Agroinfiltration method. Then, we collect apoplastic fluids from the leaves and purify the His-tagged protein by Ni2+-affinity chromatography. In addition to plant-specific post-translational modifications, protein accumulated in the plant apoplastic space, rather than in the cytosolic space, should be kept under an oxidizing environment. Such an environment will help to maintain the property of intrinsic disulfide bonds in the protein of interest. Further, purification from the apoplastic fluids, rather than the total protein extract, will significantly reduce contaminants (for instance RuBisCO) during protein extraction, and simplify downstream processes. We envisage that our system will be useful for expressing various plant proteins, particularly the apoplastic or extracellular regions of membrane proteins.
Keywords: Protein expression and purificationBackground
The use of Nicotiana benthamiana as a host organism for protein expression has increasingly become an attractive system, in addition to well-established expression systems like E. coli, yeast, insect cells, and cell-free. However, it is challenging to obtain near homogeneity protein by one-step affinity purification in the N. benthamiana system, possibly due to the complexity of the total protein extract (Souza, 2015). An alternative option is to ensure that the proteins of interest are expressed and accumulated in specialized compartments, which are spatially separated from contaminants derived from other compartments. Thus, by collecting proteins of interest accumulated in a specific compartment, we can simplify the heterogeneity of the starting material for column chromatography.
Many plant proteins passing through the secretory pathway need to get oxidized and form disulfide bonds, to mature into a stable form (Meyer et al., 2019). The appropriate disulfide bonding is required in many apoplastic proteins or extracellular regions of membrane proteins, including membrane-bound leucine-rich repeat (LRR) receptor-like kinase, and secreted peptide hormones (Meyer et al., 2019). However, the cytoplasm is normally maintained in a reducing environment. It is desirable to accumulate the expressed proteins in the apoplast, and retrieve them with appropriate disulfide bond modifications.
In this protocol, we describe a method to express and purify the ectodomain of an LRR receptor-like kinase CARD1/HPCA1 (CARD1ecto) from N. benthamiana apoplastic fluid. We will primarily focus on its expression and accumulation in the apoplast (Procedure A), its extraction from the apoplastic space (Procedure B), and its purification by Ni2+-affinity chromatography (Procedure C). This system should prove useful not only for CARD1ecto, but also for other plant proteins of interest, particularly the apoplastic or extracellular regions of membrane proteins, which may require redox-related modifications to function correctly.
Materials and Reagents
1 mL needleless plastic syringe (TERUMO, catalog number: SS-01T)
20 mL needleless plastic syringe (TERUMO, catalog number: SS-20ESz)
50 mL centrifuge tube (FALCON, catalog number: 352070)
GD/X syringe filter (PES 0.45 μm) (GE healthcare, catalog number: 6876-2504)
500 mL Pyrex beaker
300 mL Pyrex beaker
Vacuum desiccator
15 mL centrifuge tube (FALCON, catalog number: 352097)
Soil-grown Nicotiana benthamiana
pEAQ-HT plasmid (Sainsbury et al., 2009)
Agrobacterium tumefaciens C58C1 carrying pCH32 (Hamilton et al., 1996; Hellens et al., 2000)
LB Broth (Lennox) (Sigma-Aldrich, catalog number: L7275-500TAB)
Kanamycin (FUJIFILM Wako Chemicals, catalog number: 113-00343)
Rifampicin (FUJIFILM Wako Chemicals, catalog number: 185-01003)
4'-Hydroxy-3',5'-dimethoxyacetophenone (acetosyringone) (Sigma-Aldrich, catalog number: D134406-25G)
Bis-Tris (Dojindo, catalog number: 6976-37-0)
Tween 20 (polyoxyethylene sorbitan monolaurate) (Nacalai Tesque, catalog number: 35624-15)
cOmpleteTM ULTRA Tablets, EDTA-free, Protease Inhibitor Cocktail (Merck, catalog number: 5892953001)
Sodium Chloride (FUJIFILM Wako Chemicals, catalog number: 195-01663)
Magnesium Chloride (FUJIFILM Wako Chemicals, catalog number: 136-03995)
Immobilized Ni2+-affinity column, HisTrap excel (Cytiva, catalog number: 17371205)
Note: Whilst any standard Ni2+-affinity column should be acceptable, we recommend using the HisTrap excel column by Cytiva. In this method, the His-tagged protein will be purified in buffer at pH 6.0, given that apoplast space is usually weakly acidic. According to the manufacturer’s instructions, HisTrap excel can capture His-tagged proteins even at pH 6.0.
Superloop, 1/16" fittings (ÄKTAdesign), 50 mL (Cytiva, catalog number: 18111382)
Imidazole (FUJIFILM Wako Chemicals, catalog number: 095-00015)
Vivaspin Turbo 15 (10 kDa molecular weight cut off) (Sartorius, catalog number: VST15T01)
Coomassie protein stain, such as InstantBlue (Expedeon, catalog number: ISB01L)
Agroinfiltration buffer (see Recipes)
Vacuum infiltration buffer (see Recipes)
Equilibration buffer (see Recipes)
Wash buffer (see Recipes)
Elution buffer (see Recipes)
Equipment
Vacuum pump (e.g., ULVAC DTC-41, or equivalent model)
Electroporation system (e.g., Bio-Rad Gene Pulser XCellTM, or equivalent)
Centrifuge with a swing rotor for 50 mL centrifuge tubes (e.g., Hitachi CF16RXII with swing rotor T4SS31, or equivalent)
Centrifuge with a fixed angle rotor for 15 mL centrifuge tubes (e.g., Hitachi CR20GIII with fixed angle rotor R15A, or equivalent)
Fast Protein Liquid Chromatography machine (we used the equivalent of GE healthcare AKTA pure 25 M1 (Cytiva, catalog number 29018227), with optional accessories installed).
Note: Any fast protein liquid chromatography machine from companies such as Cytiva (https://www.cytivalifesciences.com/) or from Bio-Rad (https://www.bio-rad.com/) are fine.
Spectrophotometer (e.g.,Thermo Fisher Scientific NanodDrop OneC, or equivalent)
Standard SDS-PAGE equipment (such as Mini-PROTEAN® Tetra Cell from Bio-Rad). For the SDS-PAGE protocol, please refer to Green and Sambrook (2014).
Procedure
Expression of His-tagged CARD1 ectodomain in apoplastic space of N. benthamiana
Note: We expressed non-tagged CARD1ecto, and purified it by conventional column chromatographies in our previous study (Laohavisit et al., 2020). Although it is possible to purify non-tagged proteins, the purification procedure is simpler in His-tagged proteins.
Clone the nucleotide sequence encoding CARD1 from Arabidopsis genomic DNA (amino acids 1–546) into the pEAQ-HT vector, in frame with a His tag at C-terminus (CARD1ecto-His), using standard molecular biology techniques (Figure 1).
Notes:
The native signal peptide of CARD1 should be intact, so that CARD1ecto-His is secreted into the apoplastic space.
In the pEAQ-HT vector system, a gene of interest is inserted between a modified 5’-untranslated region (UTR) and the 3’-UTR from Cowpea mosaic virus RNA-2, and co-expressed together with silencing suppressor p19 (Sainsbury et al., 2009; Sainsbury and Lomonossoff, 2008). This permits an extremely high-level and rapid production of proteins of interest (see Figure 1).
We used genomic DNA to clone the construct, but cDNA should also work.
For His-tag at C-terminus, pEAQ-HT should be digested with AgeI and SmaI restriction enzymes (Figure 1, see Sainsbury et al., 2009 for details).
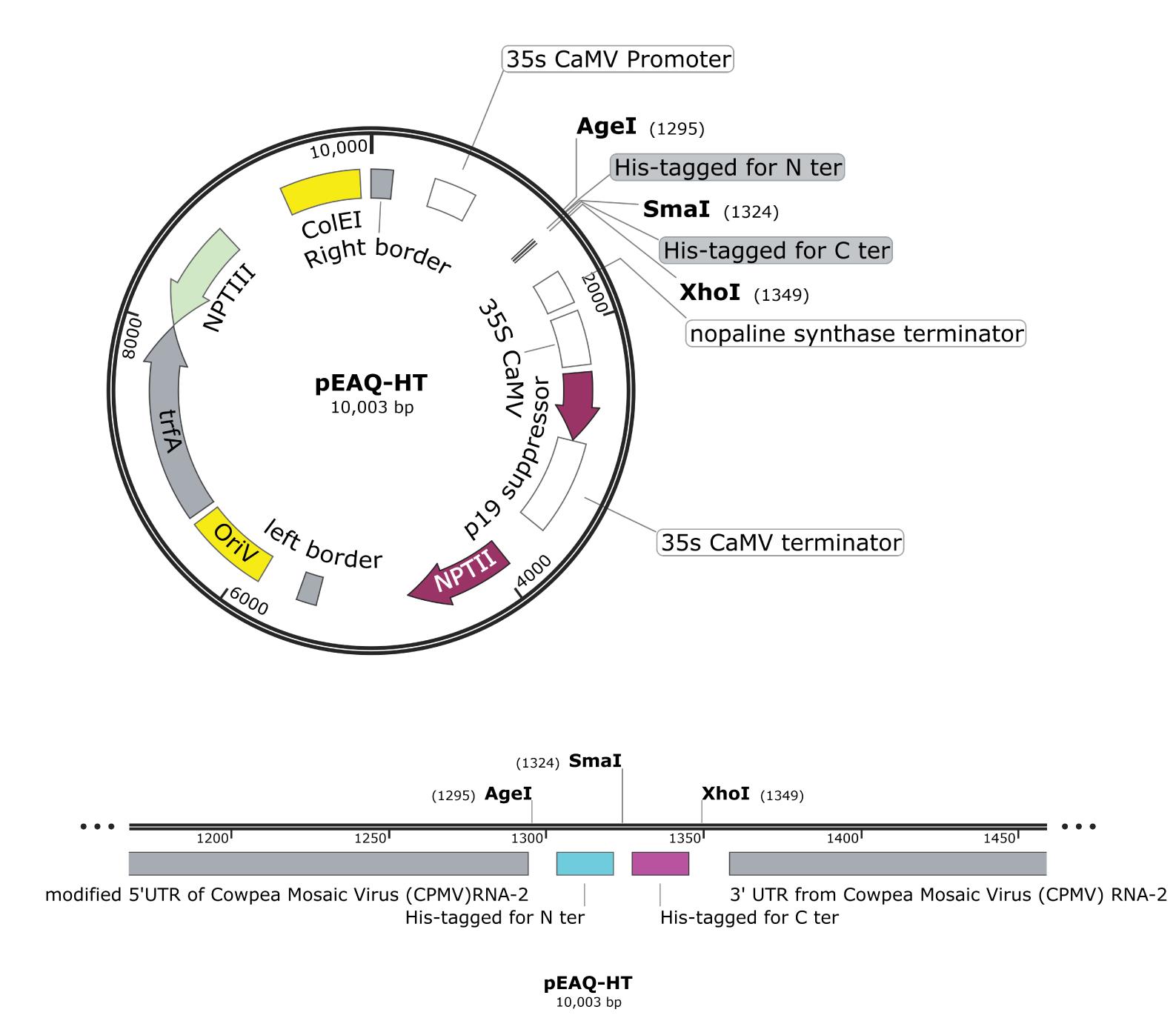
Figure 1. The pEAQ-HT vector map. Top, a simplified pEAQ-HT vector map. Bottom, a close-up of the restriction enzyme map.After sequence confirmation, transform electrocompetent cells of Agrobacterium tumefaciens C58C1 with pEAQ-CARD1ecto-His, using an electroporation system (machine default settings for Agrobacterium tumefaciens are capacitor = 25 μF, pulse controller = 200 Ω, and Voltage = 2.4 kV). Plate out the transformed cells onto LB agar plates supplemented with 100 μg/mL rifampicin and 50 μg/mL kanamycin.
Confirm the Agrobacterium colonies with correct transformation using PCR, and make glycerol stocks for long term storage.
Pick a fresh colony or take a small portion of the glycerol stocks of Agrobacterium into 5 mL of LB liquid media supplemented with 100 μg/mL rifampicin and 50 μg/mL kanamycin and incubate it with shaking at 120 rpm at 28°C overnight.
Dilute 2 mL of overnight culture with 18 mL of fresh LB liquid media supplemented with 100 μg/mL rifampicin and 50 μg/mL kanamycin, and incubate with shaking at 120 rpm and 28°C for 4 h.
Centrifuge the Agrobacterium at 3,000 × g for 5 min, and discard the supernatant.
Add 10 mL of Agroinfiltration buffer, and resuspend the pellet.
Repeat steps 6–7.
Measure OD600 of the suspension and adjust it to 0.3.
Infiltrate the Agrobacterium suspension into whole leaves of 5-weeks-old N. benthamiana from the abaxial side, using 1 mL needleless syringes. The infiltrated area should turn darker in color compared to the unfiltrated area (Yin et al., 2017).
Note: In our experiment, we infiltrated approximately 30 leaves to obtain approximately 1 mg of highly purified CARD1ecto-His protein. For other proteins of interest, it is recommended that the experimenter perform small-scale pilot experiments (for instance, 8–10 leaves per construct) to ascertain how much protein can be obtained, since different proteins will show different expression levels.
Extraction of apoplastic fluid from N. benthamiana leaves
After 4–7 days post inoculation, harvest the infiltrated N. benthamiana leaves and pile them together, such that the abaxial side of the leaves points upward.
Carefully place the pile of leaves into a 500-mL Pyrex beaker containing 150 mL of vacuum-infiltration buffer. Afterward, place a 300-mL Pyrex beaker on top of the pile, such that leave samples are submerged in buffer (Figure 2).

Figure 2. Set-up for vacuum infiltration.Place the assembled beaker (with leave samples) into a vacuum desiccator, and apply a pressure of 60 hPa for 10 min using a pump (or other appropriate equipment). Bubbles should be released from the leaves.
Slowly release the pressure inside the desiccator. We usually allow at least 10 min for the pressure to return to normal.
Note: In this step, expanded air bubbles in the apoplastic space shrink significantly and vacuum-infiltration buffer will replace this space. The color of the leaves should turn darker as the apoplastic space has been filled with buffer. If you do not notice any differences, this suggests that the vacuum infiltration process has not been successful, and step 3 should be carefully repeated.
Carefully take out individual leaves from the beaker and gently remove excess buffer using paper towels. Set these aside.
Meanwhile, assemble the apoplastic fluid collection unit. This consists of the following materials:
20-mL single-use needleless plastic syringe with the plunger removed (i.e., only the barrel part is needed)
50-mL centrifuge tubes, non-skirted
To assemble the apoplastic fluid collection unit, simply place the barrel part of a 20-mL sized syringe into a 50-mL centrifuge tube (Figure 3A).
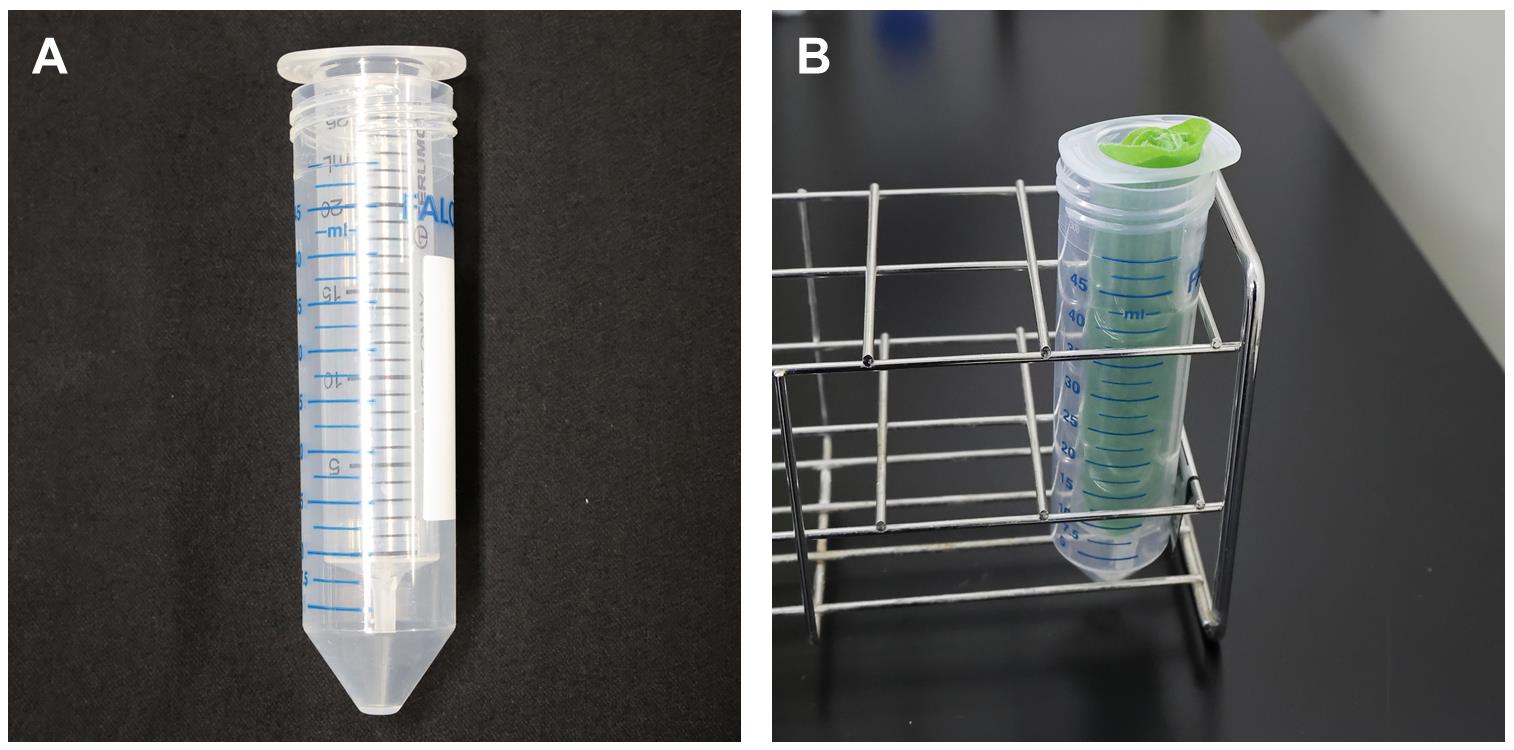
Figure 3. Set-up of the apoplastic fluid collection unit.Per apoplastic fluid collection unit, take 6–8 leaves from step 5 and separate them into two piles (3–4 leaves per pile). Next, carefully roll the leaves from one pile (from the base to the apex of the leaf) and place this into the barrel part of the apoplastic fluid collection unit. The abaxial or adaxial sides can be faced in any direction. Repeat for the second pile (Figure 3B).
Note: For our experiment, we used approximately 30 leaves to purify CARD1ecto-His protein, which corresponded to 6 apoplastic fluid collection units.
Place the apoplastic fluid collection units (with leaves) in a swing rotor, and centrifuge at 1,500 × g and 4°C for 10 min.
Note: Centrifugation should be performed using a swing rotor, to maximize recovery of the apoplast fluid.
Apoplastic fluid should appear at the bottom of the 50-mL centrifuged tubes. Transfer the fluid to a separate tube and centrifuge again at 1,500 × g and 4°C for 3 min, to ensure all fluid has been extracted. Finally, pool all the apoplastic fluid into a single tube.
Centrifuge the collected apoplastic fluid from step 9 at 10,000 × g and 4°C for 10 min. Collect the supernatant, and centrifuge it again at 10,000 × g and 4°C for 10 min.
Filter the supernatant with a GD/X syringe filter. The resulting filtrate is now ready for the next step of purification.
Note: The filter unit should have a 0.45 µm pore size.
CARD1ecto-His purification by Ni2+-affinity chromatography
Note: Monitor the absorbance at 280 nm to detect proteins throughout the purification process.
Connect a HisTrap excel column (capacity of 5 mL) to an FPLC system (such as the AKTA pure), and wash this column with 25 mL of ultrapure water (5 column volumes, CV) at 5 mL/min (1 CV/min).
Note: Wash volume and the flow rate should be adjusted according to the column size used.
Equilibrate the column with 25 mL (5 CV) of equilibration buffer at 5 mL/min (1 CV/min).
Load the apoplastic fluid sample onto the column with a superloop at 2.5 mL/min (0.5 CV/min).
Note: Depending on your FPLC machine configuration and your sample volume, you can also load your sample manually.
Wash the column with 25 mL (5 CV) of wash buffer at 5 mL/min (1 CV/min).
Elute the His-tagged protein with 25 mL (5 CV) of elution buffer at 5 mL/min (1 CV/min).
Pool the peak fractions in one sampling tube and check the purity of CARD1ecto-His by SDS-PAGE, followed by Coomassie brilliant blue staining (InstantBlue; Expedeon) (Figure 4).
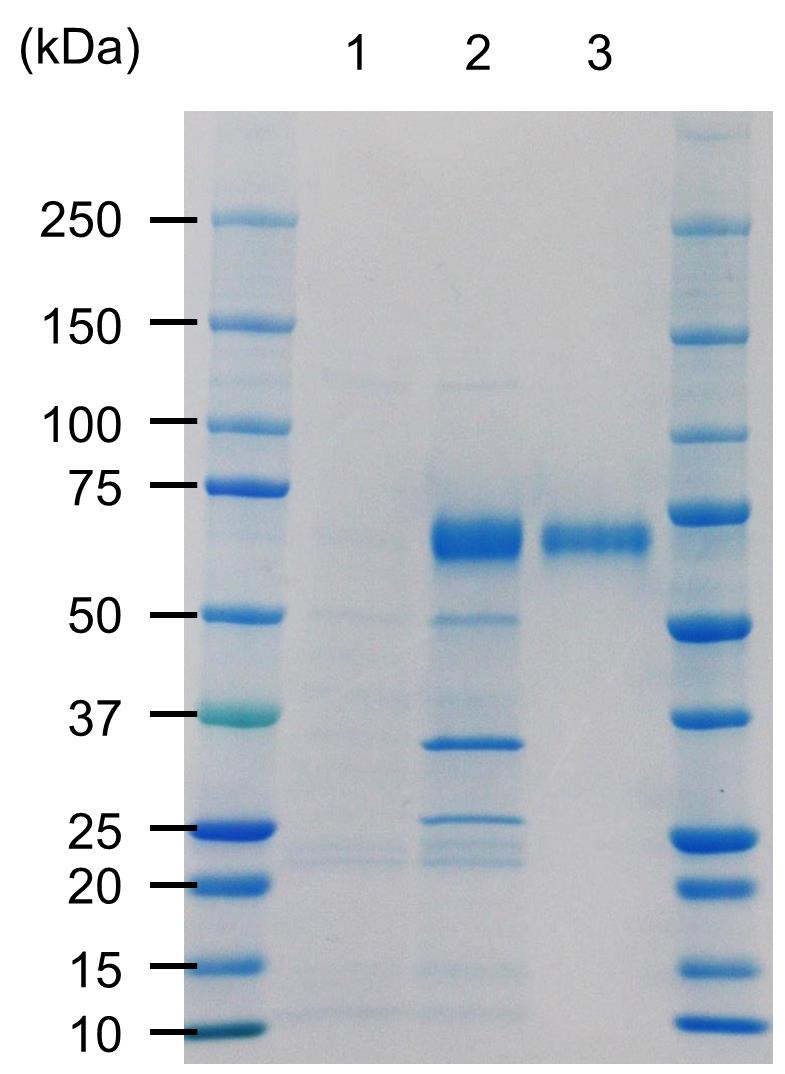
Figure 4. SDS-PAGE analysis of purified CARD1ecto-His. Lane 1: Apoplastic fluid from intact N. benthamiana leaves. Lane 2: Apoplastic fluids from the CARD1ecto-His expressing N. benthamiana leaves. Lane 3: Purified CARD1ecto-His by Ni2+-affinity chromatography. The gel was stained with Coomassie brilliant blue (InstantBlue).Measure protein concentration by absorbance at 280 nm, using a spectrophotometer (such as NanoDrop OneC).
(Optional) Apply the pooled fractions in an ultrafiltration unit with a 10-kDa cut-off membrane (Vivaspin turbo 15) for buffer exchange and/or concentration.
Note: When buffer exchanging or sample concentration are necessary for downstream processes.
Recipes
Agroinfiltration buffer
10 mM MES/NaOH (pH 5.6)
10 mM MgCl2
150 μM acetosyringone
Vacuum infiltration buffer
20 mM Bis-Tris/HCl (pH 6.0)
0.01% Tween20
1× cOmpleteTM Protease Inhibitor Cocktail
Equilibration buffer
20 mM Bis-Tris/HCl (pH 6.0)
300 mM NaCl
Wash buffer
20 mM Bis-Tris/HCl (pH 6.0)
300 mM NaCl
5 mM Imidazole
Elution buffer
20 mM Bis-Tris/HCl (pH 6.0)
300 mM NaCl
300 mM Imidazole
Acknowledgments
This protocol was adapted from our published work (Laohavisit et al., 2020). This work was supported by JSPS Grant-in-Aid for 17H06172 and 20H05909 to K.S.
Competing interests
The authors declare no competing interests.
References
- Green, M.R. and Sambrook, J. (2014). Molecular Cloning: A Laboratory Manual (4th Edition). Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press.
- Hamilton, C.M., Frary, A., Lewis, C. and Tanksley, S.D. (1996). Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci U S A 93(18): 9975-9979.
- Hellens, R., Mullineaux, P. and Klee, H. (2000). Technical Focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Science 5(10): 446-451.
- Laohavisit, A., Wakatake, T., Ishihama, N., Mulvey, H., Takizawa, K., Suzuki, T. and Shirasu, K. (2020). Quinone perception in plants via leucine-rich-repeat receptor-like kinases. Nature 587(7832): 92-97.
- Meyer, A.J., Riemer, J. and Rouhier, N. (2019). Oxidative protein folding: state-of-the-art and current avenues of research in plants. New Phytol 221(3): 1230-1246.
- Sainsbury, F. and Lomonossoff, G. P. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol 148(3): 1212-1218.
- Sainsbury, F., Thuenemann, E. C. and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7(7): 682-693.
- Souza, A. D. (2015). Expression and partial purification of His-tagged proteins in a plant system. Bio-protocol 5(17). e1572.
- Yin, K., Han, T. and Liu, Y. (2017). Use of Geminivirus for delivery of CRISPR/Cas9 components to tobacco by Agro-infiltration. Bio-protocol 7(7): e2209.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ishihama, N., Laohavisit, A., Takizawa, K. and Shirasu, K. (2022). Apoplastic Expression of CARD1-ecto Domain in Nicotiana benthamiana and Purification from the Apoplastic Fluids. Bio-protocol 12(8): e4387. DOI: 10.21769/BioProtoc.4387.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









