- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Quick Method to Quantify Iron in Arabidopsis Seedlings
(*contributed equally to this work) Published: Vol 12, Iss 5, Mar 5, 2022 DOI: 10.21769/BioProtoc.4342 Views: 3922
Reviewed by: Wenrong HeKevin RobeMin Cao

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Intact Vacuoles from Arabidopsis Root Protoplasts and Elemental Analysis
Chuanfeng Ju [...] Zhenqian Zhang
Mar 5, 2023 2038 Views

High-Performance Liquid Chromatography Quantification of Glyphosate, Aminomethylphosphonic Acid, and Ascorbate in Culture Medium and Microalgal Cells
Juan Manuel Ostera [...] Gabriela Malanga
Apr 5, 2025 1172 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 543 Views
Abstract
Iron (Fe) is an indispensable micronutrient for plant growth and development. Since both deficiency, as well as a surplus of Fe, can be detrimental to plant health, plants need to constantly tune uptake rates to maintain an optimum level of Fe. Quantification of Fe serves as an important parameter for analyzing the fitness of plants from different accessions, or mutants and transgenic lines with altered expression of specific genes. To quantify metals in plant samples, methods based on inductively coupled plasma-optical emission spectrometry (ICP-OES) or inductively coupled plasma-mass spectrometry (ICP-MS) have been widely employed. Although these methods are highly accurate, these methodologies rely on sophisticated equipment which is not always available. Moreover, ICP-OES and ICP-MS allow for surveying several metals in the same sample, which may not be necessary if only the Fe status is to be determined. Here, we outline a simple and cost-efficient protocol to quantify Fe concentrations in roots and shoots of Arabidopsis seedlings, by using a spectroscopy-based assay to quantify Fe2+-BPDS3 complexes against a set of standards. This protocol provides a fast and reproducible method to determine Fe levels in plant samples with high precision and low costs, which does not depend on expensive equipment and expertise to operate such equipment.
Keywords: ArabidopsisBackground
Iron (Fe) is an essential micronutrient, which is involved in numerous biochemical and physiological processes in plants. A deficiency or an excess of Fe in plants limit plant growth and cause severe losses in crop yield and quality. Imbalances of Fe levels in plants also affect the homeostasis of other nutrients (Schmidt et al., 2020). Therefore, the determination of Fe concentrations in plant tissues is mandatory for the assessment of the nutritional status of the plant and serves as an important parameter for studying Fe-related genes. Currently, inductively coupled plasma (ICP) spectrometry-based methods are considered the gold standard for determining Fe levels. Although these methods are highly sensitive and reliable, their accessibility is rather limited due to costly equipment, specialized training required for the operation of such equipment, and the requirement of generally large amounts of material, which may render the analysis difficult in cases where sample size is limited. Therefore, a simple, reliable, and easily accessible method to measure Fe levels at high accuracy is a valuable alternative to ICP-based methods.
Here, we describe a method for quantifying total Fe in roots and shoots of plant seedlings. The method is based on a protocol established for the analysis of Fe concentration in Plantago lanceolata L. leaves (Schmidt, 1996), which we have adapted and optimized for convenient and routine analysis of the more commonly used model plant Arabidopsis thaliana. The method involves wet acid digestion of dried plant samples with nitric acid and hydrogen peroxide, to solubilize Fe and reduction of Fe(III) complexes by hydroxylammonium chloride, followed by a colorimetric assay based on the ability of the chelating agent bathophenanthroline disulfonate (BPDS) to form red-colored complexes with ferrous Fe. The resulting Fe2+(BPDS)3 complex has a distinct absorbance band at 535nm under acid conditions, allowing for the quantification of Fe concentrations by interpolation from a standard curve. BPDS has been widely used for the determination of Fe levels in a variety of samples (Pré and Benlatrèche, 1977; Tangerås, 1983; Hirayama and Nagasawa, 2017; Freinbichler et al., 2020). The present protocol was optimized for Arabidopsis thaliana samples, and has been used for quantifying Fe concentrations as low as 50 μg/g dry weight sample or less (Tsai et al., 2018; Gautam et al., 2021), but can be modified and applied to determine Fe levels in samples from other plant species.
Materials and Reagents
Butter paper
Tissue paper/Kimwipes
15 mL screw cap conical tubes (Sarstedt, catalog number: NC1377856)
96-well microplate (Greiner CELLSTAR®, catalog number: 655180)
Nitric acid (65%) (Merck, catalog number: 1.00456.1000, store at room temperature)
Hydrogen peroxide solution (30%) (Merck, catalog number: 1.07209.1000, store at room temperature)
ddH2O
Bathophenanthrolinedisulfonic acid disodium salt hydrate (Sigma, catalog number: B1375-5G, store at room temperature)
Sodium acetate (Sigma, catalog number: S8750-500G, store at room temperature)
Hydroxylamine hydrochloride (Alfa Aesar, catalog number: A15398.36, store at room temperature)
Iron (III) chloride hexahydrate (Merck, catalog number: 1.03943.0250, store at room temperature inside a dry cabinet)
Assay solution (see Recipes)
Fe standard solutions (see Recipes)
Equipment
Vannas scissors
Drying oven (Hot air oven, PREMA®)
Vortex mixer (Vortex-Genie 2, Scientific Industries)
Heat block (Elite Dry Bath Incubator, Major Science)
Microplate spectrophotometer (Power Wave XS2, BioTek Instruments, Agilent Technologies)
Software
Gen5TM BioTek Instruments (Agilent Technologies)
GraphPad Prism version 9
Procedure
Sampling
Sow seeds on Estelle and Somerville (ES) nutrient media (Estelle and Somerville, 1987), and stratify for at least two days in the dark at 4°C, before transferring to a growth chamber. Grow seedlings at 21–22°C under continuous illumination (50 μmol m2 s−1) for two weeks.
To separate seedlings into shoots and roots for sample harvesting, use Vannas scissors to cut seedlings on the media at hypocotyl junctions.
For shoot samples, collect 25–30 shoots of two-week-old seedlings in butter paper.
For root samples, collect 30–36 roots of two-week-old seedlings in butter paper. Before placing roots into butter paper, make sure that the roots don’t contain any residual media, which also contains Fe. Remove any media from the roots with tissue paper/Kimwipes, followed by washing twice with ddH2O. Then gently dry roots with tissue paper/Kimwipes.
Dry samples in an oven for at least two days at 60–65°C.
Determine the dry weight of each sample.
After weighing, transfer the samples into 15 mL conical tubes.
Sample digestion
Add 225 µL of nitric acid (65%) into each tube and screw the caps tightly.
Incubate for 6 h at 95°C on a heat block, vortexing the tubes every 1–2 h. Make sure that samples do not stick to the walls of the tubes and no precipitates are present, as these could affect measurements.
Carefully unscrew the caps of the tubes, add 150 µL of H2O2 (30%) into each tube, and screw the caps back tightly.
Incubate for 2 h at 56°C on a heat block, and vortex every 30–60 min to make sure no precipitates are formed, as this could also affect measurements.
Add 225 µL of ddH2O. Fully digested samples should be pale yellow in color, and ideally should have no white precipitates.
For the ease of pipetting in the following steps, carefully transfer the digested samples into 1.5 mL microcentrifuge tubes. At this point, samples are ready to be immediately used for the Fe quantification in procedure C, or can be stored at 4°C in the dark for up to 1 month for later analysis.
Standard and sample preparation, and spectroscopic reading
Prepare the assay solution (see Recipe 1) and Fe standard solutions (see Recipe 2).
Add 5 µL of standards/samples in microplate wells. Run three technical replicates for each standard/sample.
Add 245 µL of assay solution to each well with standard/sample.
Cover the microplate and keep it in the dark to incubate at room temperature for 5 min before spectrophotometric measurement. Following incubation, the standards will turn pink/red, with increasing color saturation as the Fe level increases (Figure 1A), and the samples will turn light yellowish-pink/orange-red depending on the Fe content.
Measure absorbance at 535 nm. Use the 0 Fe standards as blanks to zero the plate reader.
Data analysis
Create a standard curve in an Excel sheet by plotting the average blank-corrected 535 nm absorbance values vs. the concentration of standards. Make sure that the coefficient of determination (R2) value is ≥0.990.
Use the equation derived from the standard curve to calculate the Fe concentrations in the samples. Table 1 shows the calculation of Fe concentrations in root samples.
Divide the Fe concentrations of the samples by their respective dry weight (DW), to obtain the final Fe concentration per g DW (Table 1).
Plot the final graph in Excel or any suitable software. The representative plot shown in Figure 1B was generated using GraphPad Prism 9. The sample data shown only represents one biological replicate, with three technical replicates. A complete experiment should have at least three biological replicates.
Table 1. Calculations to quantify Fe in root samples.
Spectrophotometric readings of the standards were used to plot a calibration curve. The Fe concentrations in the samples were calculated using the equation derived from the slope. Wild-type (Col-0) plants, a coumarin-deficient mutant (f6’h1-1; Schmid et al., 2014), and an Fe over-accumulating genotype (IMA2ox; Gautam et al., 2021) grown on nutrient media containing 50 µM Fe-EDTA are used to represent Fe quantification. The data represent the standard deviation of three technical replicates from the spectrophotometer reading. The microplate layout used to make spectrophotometric readings is shown at the top. STD, standard; S, sample; Abs., absorbance; DW, dry weight; Avg., average; SD, standard deviation.
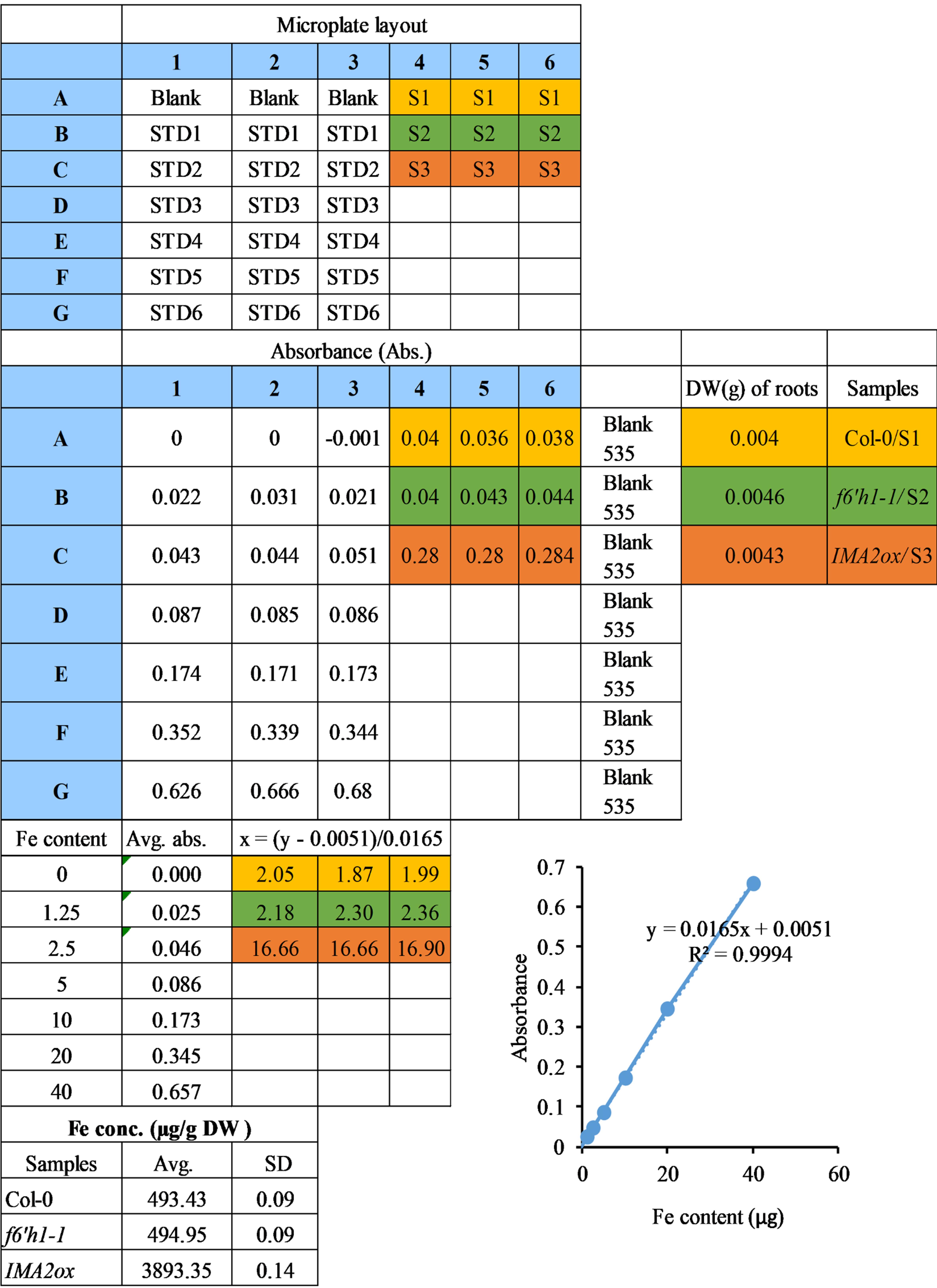
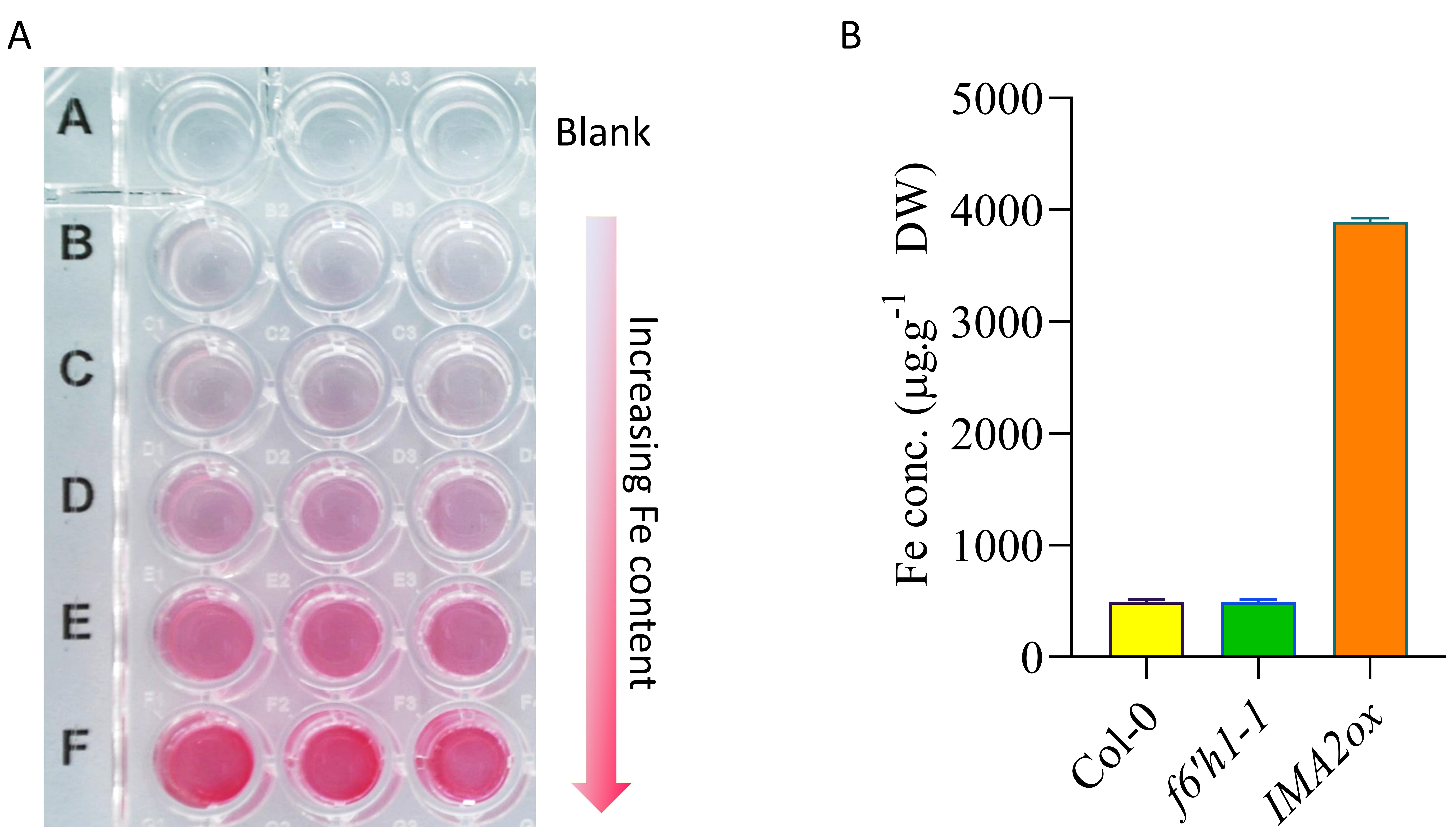
Figure 1. Fe content-dependent color pattern of Fe2+(BPDS)3 complexes and their spectrophotometric quantification. A. Representative figure showing the change in color of Fe2+(BPDS)3 complex across an increasing Fe content gradient. Equal volume of assay solution was added in a 96 well plate microplate containing different amounts of Fe (0, 2.5, 5, 10, 20, and 40 µg) in three technical repeats. The microplate was kept in the dark to incubate at room temperature for 5 min, a photo was captured immediately afterwards. B. Graph showing the Fe concentrations in different samples (genotypes). Wild-type (Col-0) plants, a coumarin-deficient mutant (f6’h1-1; Schmid et al., 2014), and an Fe over-accumulating genotype (IMA2ox) (Gautam et al., 2021) grown on nutrient media containing 50 µM of Fe-EDTA are used to represent the Fe quantification. The data represent standard deviation of three technical replicates from the spectrophotometer reading. DW, dry weight.
Notes
This method can be used to quantify Fe in samples grown on both soil and nutrient media.
Steps B to C need to be carried out inside a fume hood with proper protection, including corrosion resistant gloves, safety glasses, lab coat, and shoes.
The sample volume should be reduced or the reagent volumes should be adjusted for complete digestion of samples.
Use only high-grade acid-resistant falcon, PTFE, or TeflonTM tubes for acid digestion. Make sure to tightly cap the tubes during the wet acid digestion steps.
Although the volume of the digested sample is only 600 µL, a 15 mL Falcon tube is used for safety reasons, and to avoid damaging the tubes due to pressure from gas build-up during the heat assisted digestion steps.
Recipes
Assay solution (for 100 mL)
Note: Prepare fresh assay solution in a light-shield bottle and store at 4°C until use.No. Reagent Working concentration Amount to add
1.Bathophenanthrolinedisulfonic acid disodium salt hydrate (MW: 590.53
g/mol)1 mM 0.06 g 2. Sodium acetate (MW: 82.03 g/mol) 0.6 M 4.92 g
3.Hydroxylamine
hydrochloride (MW: 69.49 g/mol)0.48 M 3.34 g 4. ddH2O - adjust to 100 mL Fe standard solutions
Prepare a 50 mM FeCl3 stock solution (50 mM FeCl3 contains 2.79 µg/µL of Fe). With this stock solution, prepare Fe standards in 1.5 mL microcentrifuge tubes as follows:
Standards Blank STD1 STD2 STD3 STD4 STD5 STD6 Fe content (µg) 0 1.25 2.5 5.0 10.0 20.0 40.0 Amount of stock to add (µL) 0 0.45 0.90 1.79 3.58 7.17 14.34 Nitric acid (65%) 225 µL into each tube H2O2 (30%) 150 µL into each tube ddH2O 225 µL into each tube
Acknowledgments
This work was supported by a grant from the Ministry of Science and Technology to W.S. (grant No.: 108-2311-B-001-033-MY3). The data used to create the representative graph and table has been extracted from previously published work (Gautam et al., 2021).
Competing interests
The authors declare no conflict of interest.
References
- Freinbichler, W., Misini, B., Colivicchi, M. A., Linert, W., Tipton, K. F. and Della Corte, L. (2020). The application of bathophenanthroline for the determination of free iron in parallel with hROS in microdialysis samples. J Neurosci Methods 331: 108530.
- Gautam, C. K., Tsai, H. H. and Schmidt, W. (2021). IRONMAN tunes responses to iron deficiency in concert with environmental pH. Plant Physiol 187(3): 1728-1745.
- Hirayama, T. and Nagasawa, H. (2017). Chemical tools for detecting Fe ions. J Clin Biochem Nutr 60(1): 39-48.
- Pré, J. and Benlatrèche, C. (1977). Rapid determination of serum iron concentration using bathophenanthroline sulfonate in a formate buffered system. Pathol Biol(Paris) 25(3): 203-205.
- Estelle, M. A. and Somerville, C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. MGG Mol Gen Genet 206: 200-206.
- Schmid, N. B., Giehl, R. F., Doll, S., Mock, H. P., Strehmel, N., Scheel, D., Kong, X., Hider, R. C. and von Wiren, N. (2014). Feruloyl-CoA 6'-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol 164(1): 160-172.
- Schmidt, W., (1996). Influence of chromium(lll) on root-associated Fe(lll) reductase in Plantago lanceolata L. J Exp Bot 47: 805-810.
- Schmidt, W., Thomine, S. and Buckhout, T. J. (2020). Editorial: Iron Nutrition and Interactions in Plants. Front Plant Sci 10: 1670.
- Tangerås, A., (1983). Iron content and degree of lipid peroxidation in liver mitochondria isolated from iron-loaded rats. Biochim Biophys Acta 757(1): 59-68.
- Tsai, H. H., Rodriguez-Celma, J., Lan, P., Wu, Y. C., Velez-Bermudez, I. C. and Schmidt, W. (2018). Scopoletin 8-hydroxylase-mediated fraxetin production is crucial for iron mobilization. Plant Physiol 177(1): 194-207.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Gautam, C. K., Tsai, H. H. and Schmidt, W. (2022). A Quick Method to Quantify Iron in Arabidopsis Seedlings. Bio-protocol 12(5): e4342. DOI: 10.21769/BioProtoc.4342.
- Gautam, C. K., Tsai, H. H. and Schmidt, W. (2021). IRONMAN tunes responses to iron deficiency in concert with environmental pH. Plant Physiol 187(3): 1728-1745.
Category
Plant Science > Plant physiology > Nutrition
Environmental science > Plant
Plant Science > Plant biochemistry > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









