- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rapid Lipid Quantification in Caenorhabditis elegans by Oil Red O and Nile Red Staining
(*contributed equally to this work) Published: Vol 12, Iss 5, Mar 5, 2022 DOI: 10.21769/BioProtoc.4340 Views: 8394
Reviewed by: Manish ChamoliAnupama SinghAnnmary Paul ErinjeriAnand Ramesh Patwardhan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipidomic Analysis of Caenorhabditis elegans Embryos
Hung-Chi Yang [...] Daniel Tsun-Yee Chiu
Sep 20, 2017 11354 Views

Total Triglyceride Quantification in Caenorhabditis elegans
Anjali Sandhu and Varsha Singh
Nov 20, 2020 5215 Views

Oil Red O Staining for Lipid Content in Caenorhabditis elegans
Feng-Yung Wang and Tsui-Ting Ching
Aug 20, 2021 7632 Views
Abstract
The ability to stain lipid stores in vivo allows for the facile assessment of metabolic status in individuals of a population following genetic and environmental manipulation or pharmacological treatment. In the animal model Caenorhabditis elegans, lipids are stored in and mobilized from intracellular lipid droplets in the intestinal and hypodermal tissues. The abundance, size, and distribution of these lipids can be readily assessed by two staining methods for neutral lipids: Oil Red O (ORO) and Nile Red (NR). ORO and NR can be used to quantitatively measure lipid droplet abundance, while ORO can also define tissue distribution and lipid droplet size. C. elegans are a useful animal model in studying pathways relating to aging, fat storage, and metabolism, as their transparent nature allows for easy microscopic assessment of lipid droplets. This is done by fixation and permeabilization, staining with NR or ORO, image capture on a microscope, and computational identification and quantification of lipid droplets in individuals within a cohort. To ensure reproducibility in lipid measurements, we provide a detailed protocol to measure intracellular lipid dynamics in C. elegans.
Graphic abstract:

Flow chart depicting the preparation of C. elegans for fat staining protocols.
Background
In Caenorhabditis elegans, lipid homeostasis is one of the many cellular processes that declines with age (Lynn et al., 2015; Johnson and Stolzing, 2019, Hammerquist et al., 2021). C. elegans are a popular model organism for many reasons, such as rapid generation time, short lifespan, determinate cell fate, and the availability of a fully sequenced genome, in addition to the myriad genetic and molecular tools that have been developed and optimized for use in this system. One such area in which C. elegans are particularly useful is the study of lipid metabolism, because of the multiple techniques that have been developed to determine lipid content in worms (Zhang et al., 2013). Analysis of specific lipid species can be conducted with the use of high-performance liquid chromatography-mass spectrometry and gas-chromatography-mass spectrometry (Castro et al., 2012; Pino et al., 2013). Although useful in determining lipid extract complexity, these methods require time for analysis and expensive machinery that isn’t readily available for all laboratories. Here, we describe two low-cost methods that take advantage of the transparency of the worm for visualizing the amount and tissue distribution of intracellular neutral lipids and require no more specialized equipment than present in most laboratories equipped to study C. elegans. C. elegans can be stained with many lipophilic dyes; two of the most readily available, easy-to-use, and biochemically validated compounds are Oil Red O (ORO) and Nile Red (NR) (Pino et al., 2013; Pang and Curran, 2014; Lynn et al., 2015; Webster et al., 2017; Stuhr and Curran, 2020). NR or 9-diethylamino-5H-benzo[α]phenoxazine-5-one is a lipophilic dye that stains intracellular neutral lipid droplets (Greenspan et al., 1985). NR allows for the quantification of lipid abundance due to the emission of green light following excitation. ORO is a fat-soluble dye that stains neutral lipids a bright red color, allowing for qualitative assessment of lipid droplet size and lipid distribution (O'Rourke et al., 2009). Staining C. elegans with these two different dyes allows for the determination of alterations in quantity and distribution of neutral lipids.
Materials and Reagents
Sterile 6 cm Petri dishes (VWR, catalog number: 25373-085)
Platinum wire (Tritech Research, catalog number: PT-9010)
200 μL micropipette tips (Genesee Scientific, catalog number: 24-151RL)
1,000 μL micropipette tips (Genesee Scientific, catalog number: 23-165R)
15 mL centrifuge tubes with EZ flip cap (ThermoFisher Scientific, catalog number: 362694)
1.5 mL microtube (Axygen Scientific, catalog number: MCT-150-C)
Parafilm (Sigma-Aldrich, catalog number: P7543)
10 mL disposable syringe (VWR, catalog number: 76290-382)
0.2 μm syringe filters (VWR, catalog number: 28145-477)
Eyelash brush (consisting of a human eyelash attached to a Pasteur pipette, generic, with tape, generic)
Microscope slides (VWR, catalog number: 48312-004)
Microscope slide cover slips (VWR, catalog number: 48366-227)
Razor blade, generic
LB powder (Teknova, catalog number: L9315)
Magnesium sulfate (MgSO4) (Sigma-Aldrich, catalog number: M2773)
Cholesterol (Sigma-Aldrich, catalog number: C8667)
Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C3881)
Streptomycin sulfate (Sigma-Aldrich, catalog number: S6501)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: 02-004-047)
Bacto agar (BD, catalog number: 214040)
Peptone (BD, catalog number: 211820)
50% sodium hydroxide (Fisher Scientific, catalog number: SS254-500)
Clorox® disinfecting bleach with CLOROMAX®–Concentrated Formula (Clorox)
Oil Red O (Alfa Aesar, catalog number: A12989)
DAPI (Sigma-Aldrich, catalog number: D9542)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650-5X10ML)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S9763)
Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P5504)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P0662)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P3911)
Triton X-100 (Sigma-Aldrich, catalog number: X100-100 ml)
Isopropanol (BHD, catalog number: BDH1133)
Ethanol (VWR, catalog number: 89125-172)
Nail polish (clear, generic)
Nile Red for microscopy (Sigma-Aldrich, catalog number: 72485)
Acetone 99.5% (BDH, catalog number: 1101-1LP)
Nematode growth medium (NGM) plates (see Recipes)
M9 (see Recipes)
Oil Red O stock solution (see Recipes)
DAPI stock solution (see Recipes)
Phosphate buffered saline (PBS) (see Recipes)
PBS + 0.01% Triton X-100 (PBST) (see Recipes)
Nile Red stock solution (see Recipes)
Worm strains (available from Caenorhabditis Genetics Center)
E. coli OP50-1 (available from Caenorhabditis Genetics Center)
Equipment
2–20 μL micropipette (Gilson, catalog numer: FA10003M)
20–200 μL micropipette (Gilson, catalog numer: FA10005M)
100–1,000 μL micropipette (Gilson, catalog numer: FA10006M)
Microcentrifuge for 1.5 mL tubes (Eppendorf, model: 5430)
Centrifuge for 15 mL tubes (Eppendorf, model: 5702)
Safe Aspiration Station (Gilson, model: F110745)
Worm incubator (Generic, maintain at 20°C)
Tube rotator (Thermo Scientific, catalog number: 88881001)
Compound microscope with DIC, DAPI, and GFP filters, and 5× and 10× objectives (Zeiss, model: AxioScope5)
Color camera (Zeiss AxioCam MRm)
Digital camera (Zeiss AxioCam ERc5s)
Stir plates, generic
Magnetic stir bars, generic
Software
Software for image acquisition
Dependent on the microscope used (in this case, we used a Zeiss Axioscope and the associated ZEN imaging software).
ImageJ (available from NIH) for image quantification
Data analysis software
We used GraphPad prism, although any software capable of t-test will suffice. ANOVA may be helpful if comparing multiple conditions, but not strictly necessary. For a pairwise comparison, a t-test can be conducted, while three or more comparisons will require an ANOVA.
Procedure
Prepare hermaphrodite worms to use for staining
Maintain the growth of strains needed for ORO fat staining and/or NR fat staining on NGM plates seeded with OP50. Allow populations to be unstarved for over three generations to avoid changes due to epigenetics.
Ramp up worm strains that will be used for fat staining (NR and/or ORO).
Note: Ramping up worms requires at least 4–6 cm plates of adult worms (around 400 worms).
Synchronize populations by alkaline hypochlorite treatment.
Prepare the bleach solution (6 mL/strain is needed).
For 6 mL of bleach solution, add 4.5 mL of autoclaved H2O, 360 μL of 50% NaOH, and 1.2 mL of bleach.
Wash adult worms off plates into 15 mL conical tubes with M9. Fill the tubes to 15 mL.
Spin down at 630 × g for 30 s.
Aspirate supernatant down to 3 mL, making sure not to disturb the pellet of worms.
Add 3 mL of bleach solution and shake vigorously for 1 min and 30 s. (A 15 mL tube will have 3 mL of bleach solution and 3 mL of M9 with worms, for a total volume of 6 mL.)
After shaking, add 9 mL of M9 and shake before spinning down at 630 × g for 30 s.
Aspirate supernatant down to the pellet. Add M9 to the top of the tube, shake, and centrifuge at 630 × g for 30 s.
Aspirate supernatant down to 3 mL, making sure not to disturb the pellet.
Add 3 mL of bleach solution and shake vigorously for 45 s or until the corpses are dissolved.
Note: This step should not exceed 1 min and 20 s, to avoid over-bleaching samples.
After shaking, add 9 mL of M9 and shake before spinning down at 630 × g for 30 s.
Aspirate supernatant down to 1 mL. Add 14 mL of M9 and spin down at 630 × g for 30 s. Repeat this wash step two to three more times.
After the last wash step, aspirate down to 1 mL. Add 7 mL of M9 and rotate samples overnight in an incubator.
Note: It is important to leave space in the tube for air, otherwise the samples will not hatch overnight. The ideal overnight incubation is 12–16 h.
Drop larval stage 1 worms onto freshly seeded 6 cm NGM plates. Aim for 100 worms per plate.
L4-larvae can be stained 48 h after dropping worms and day 3-adults can be stained 120 h after dropping worms (unless strains are developmentally delayed).
Note: Staining can also be conducted on day 1-adults (72 h after dropping the worms) and/or day 2-adults (96 h after dropping the worms).
If imaging adults, transfer worms to a fresh plate every day until the day of staining.
Note: If staining worms older than day 3 of adulthood, move worms to a fresh plate every day until they have ceased reproduction.
ORO fat staining
Prepare 60% ORO staining solution.
Make up the staining solution: Dilute ORO stock solution in autoclaved water (600 μL of ORO stock solution per 400 μL of water). Depending on the volume, prepare in either a 1.5 mL Eppendorf tube or a 15 mL centrifuge tube.
Note: 600 μL is used per sample. Make more than necessary to account for volume lost during filtration (Step B.1.d.).
Wrap the top of the tube with parafilm.
Place ORO staining solution on a tube rotator overnight.
Note: Although preparing the stain overnight is preferred, stain can be prepared up to two hours before being used in this protocol.
The next day, filter the ORO staining solution using a 10 mL plastic syringe attached to a 0.2 μm syringe filter into a new centrifuge tube.
Optional: Co-staining samples with DAPI will aid in the identification of tissues (germline versus intestine) because of nuclear size and density differences. Dilute 1 mg/mL DAPI stock solution to 10 μg/mL in the ORO staining solution (10 μL of DAPI per 1 mL of ORO staining solution).
Stain C. elegans with ORO staining solution.
Wash off synchronized worm populations from 6 cm Petri dishes with NGM seeded with E. coli OP50-1 with PBS + 0.01% Triton X-100 (PBST) into a 1.5 mL tube. For each strain, you will need ~100 worms.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL (use marks on the tube as a guide).
Wash once or twice more with PBST.
Note: These washes remove E. coli from the worms to be stained. Wash until the supernatant is clear and not cloudy with bacteria.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Add 600 μL of 60% isopropanol and rotate for 3 min at room temperature to fix worms.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Add 600 μL of 60% ORO staining solution and incubate for 2 h while rotating at room temperature.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Add 600 μL of PBST and incubate for 30 min while rotating at room temperature.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Using a cut pipette tip (to broaden the opening), pipette 14 μL of worms in PBST onto a microscope slide. If you want to line worms up, do so with the eyelash brush before covering with a cover slip.
Cover with a cover slip and seal the edges with clear nail polish. Allow to dry for 5-10 min.
Image at 10× with DIC and DAPI filters using the Zeiss AxioCam ERc5s. Representative images are shown in Figure 1. Image at least 100 worms per strain per replicate for qualitative analysis of non-Asdf versus Asdf (age-dependent somatic depletion of fat).
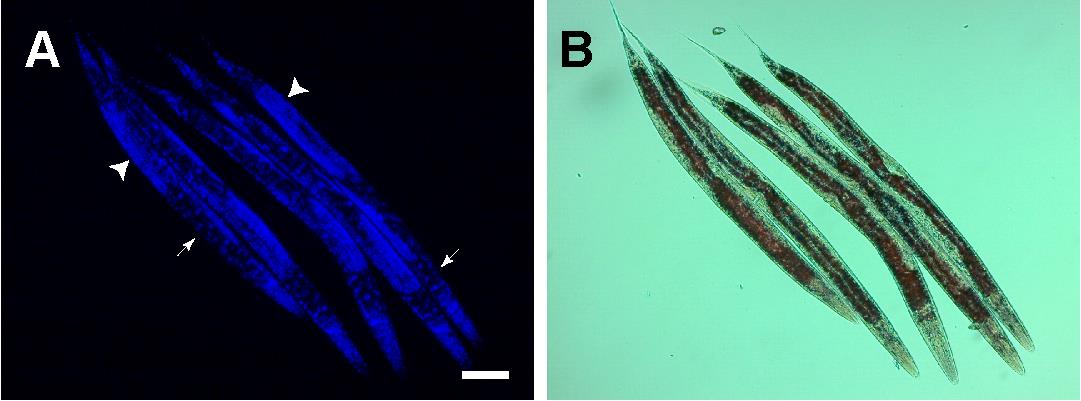
Figure 1. Representative images of skn-1gf mutant animals fixed and stained at larval stage 4 (A-B) and visualized by DAPI-stained nuclei (A) and ORO-stained lipids (B). Scale bar = 50 μm. Arrows indicate intestinal tissue; arrowheads indicate germline.
Nile Red fat staining of post-embryonic animals
Wash off synchronized worm populations from 6 cm Petri dishes with NGM seeded with E. coli OP50-1 with PBS + 0.01% Triton X-100 (PBST) into a 1.5 mL tube. For each strain, you will need ~100 worms.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL (use marks on the tube as a guide).
Wash once or twice more with PBST.
Note: These washes remove E. coli from the worms to be stained. Wash until the supernatant is clear and not cloudy with bacteria.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Add 600 μL of 40% isopropanol and rotate for 3 min at room temperature to fix worms.
While worms are fixing, make NR staining solution: dilute 0.5 mg/mL NR stock solution in 40% isopropanol (6 μL of NR stock solution per 1 mL of 40% isopropanol).
Optional: Co-staining samples with DAPI will aid in the identification of tissues (germline versus intestine). Add 1 mg/mL DAPI stock solution (10 μL in 1 mL NR staining solution).
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Add 600 μL of NR staining solution and incubate for 2 h in the dark.
Note: Before incubation, flick the bottom of the tube to resuspend the pellet in the staining solution.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Add 600 μL of PBST and incubate for 30 min in the dark.
Centrifuge for 1 min at 25 × g. Remove supernatant to 0.1 mL.
Using a cut pipette tip (to broaden the opening), pipette 14 μL of worms in PBST onto the microscope slide.
Note: If you want to line the worms up, do so with the eyelash brush before covering with a cover slip.
Cover with a cover slip and seal the edges with clear nail polish. Allow to dry in the dark for 5-10 min.
Image at 10× with DIC, DAPI (Blue), and AlexaFluor488 (Green) filters using the Zeiss AxioCam MRm. Representative images are shown in Figure 2.
Image at least 50 worms per strain per replicate for quantitative analysis.
Note: The NR fat staining is imaged in the GFP/AlexaFluor 488 channel and the DAPI is imaged in the DAPI channel.
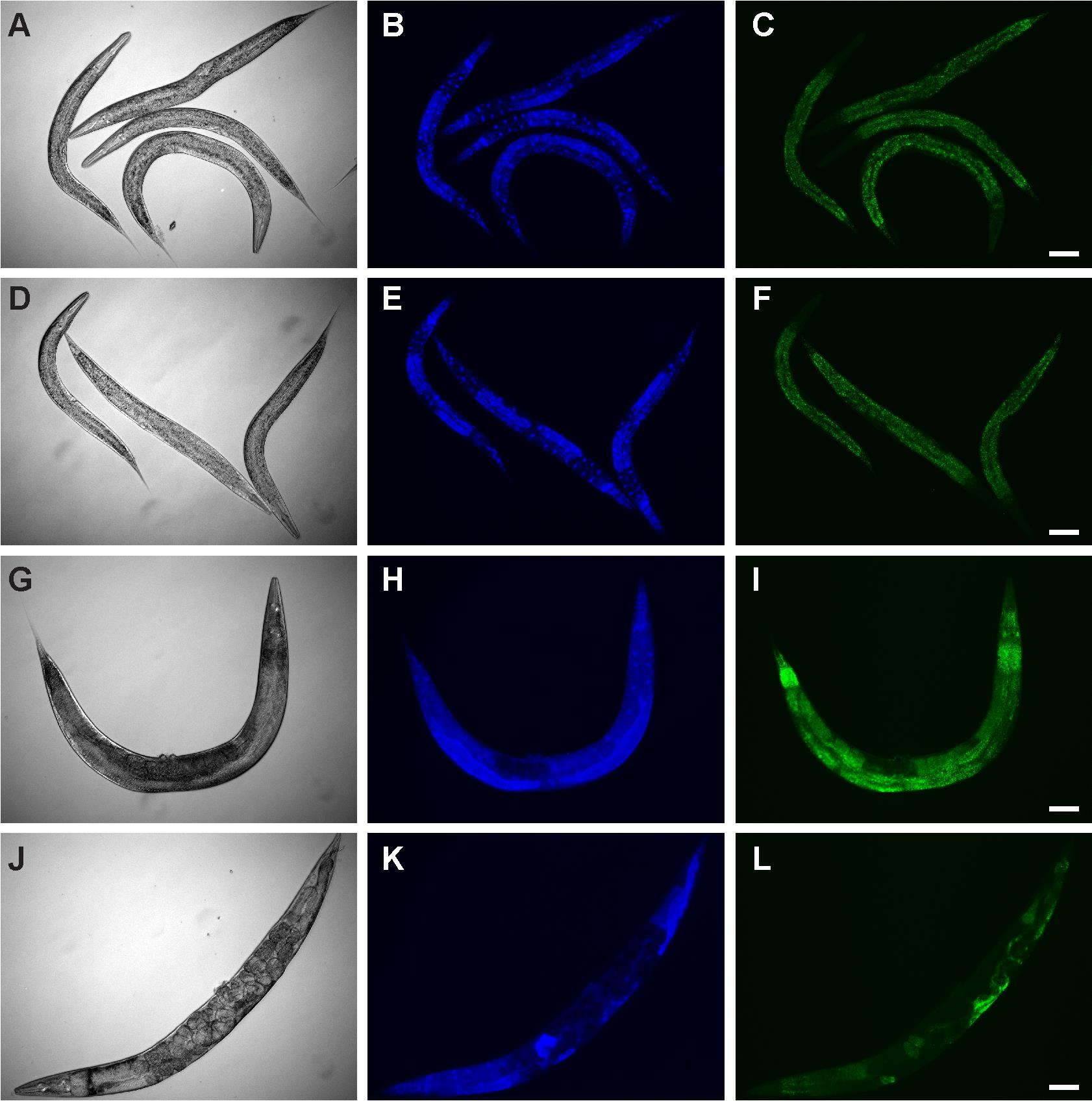
Figure 2. Representative images of wildtype (N2 Bristol) (A-C and G-I) and SKN-1gf mutant animals (D-F and J-L), fixed and stained at larval stage 4 (A-F) or day 3 of adulthood (G-L), and visualized by DIC (A, D, G, J), DAPI-stained nuclei (B, E, H, K), and NR-stained lipids (C, F, I, L). Scale bars = 50 μm.
NR fat staining of embryonic animals
Note: This protocol is similar to the NR fat staining of post-embryonic animals with a few minor differences. The minor differences include fixing embryos overnight, spinning the embryos down at 10× speed, and imaging at 40–63× magnification.
Synchronize populations by alkaline hypochlorite treatment. Instead of allowing animals to hatch overnight, aspirate down to 1 mL and transfer M9 buffer with embryos to a 1.5 mL centrifuge tube.
Spin down for 1 min at 2,500 × g. Remove as much as the supernatant as possible without disrupting the pellet.
Wash sample with PBST. Spin down for 1 min at 2,500 × g and remove supernatant to 0.1 mL.
Add 600 μL of 40% isopropanol and rotate overnight at room temperature to fix embryos and permeabilize the eggshell.
The next day, make NR staining solution: dilute NR stock solution in 40% isopropanol (6 μL of NR stock solution per 1 mL 40% isopropanol). Add 1 mg/mL of the DAPI stock solution (10 μL in 1 mL of NR staining solution).
Centrifuge for 1 min at 2,500 × g. Remove supernatant to 0.1 mL.
Add 600 μL of NR staining solution and incubate for 2 h in the dark.
Note: Before incubation, flick the bottom of the tube to resuspend the pellet in the staining solution.
Centrifuge for 1 min at 2,500 × g. Remove supernatant to 0.1 mL.
Add 600 μL of PBST and incubate for 30 min in the dark.
Centrifuge for 1 min at 2,500 × g. Remove supernatant to 0.1 mL.
Pipette 14 μL of embryos in PBST onto microscope slide.
Cover with a cover slip and seal the edges with clear nail polish. Allow to dry in the dark for 5-10 min.
Image at 40-63× with DIC, DAPI, and GFP (AlexaFluor488—Green) filters using the Zeiss AxioCam MRm. Representative images are shown in Figure 3.
Image at least 50 eggs per strain per replicate for quantitative analysis.
Note: The NR fat staining is imaged in the GFP/AlexaFluor 488 channel and the DAPI is imaged in the DAPI channel.
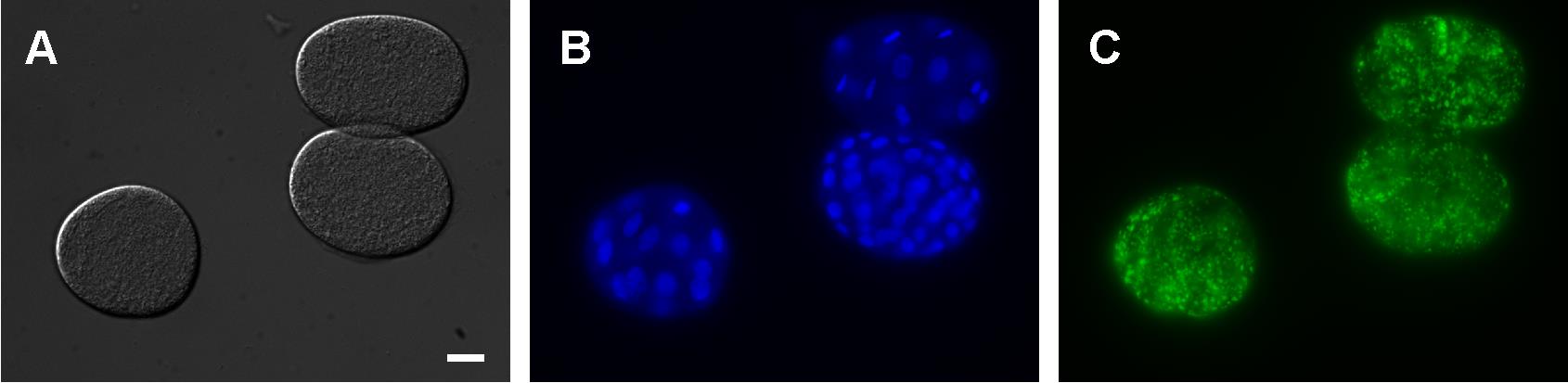
Figure 3. Representative images of fixed and stained wildtype (N2 Bristol) embryos visualized by DIC (A), DAPI-stained nuclei (B), and NR fat staining (C). Scale bar = 10 μm.
Data analysis
Quantitative analysis of NR-stained animals and embryos
Download Fiji by ImageJ to open and analyze the .czi files from the Zen imaging software: https://imagej.net/Fiji/Downloads.
Open Fiji. Open the .czi image file to analyze (File – Open – Select File – Open).
The “Bio-Formats Import Options” window will pop up. Format the following way:
For stacking view, “View stack with: Hyperstack”.
For color options, “Color mode: Custom”.
No other options need to be selected.
Once the stacking view and color options have been updated, press “OK”.
The “Console” window will pop up. You can ignore this window.
The “Bio-Formats Color Customization” window will pop up. Select a color for each channel used to image.
Example:
DIC
1) “Series 0 Channel 0 Red”: 255
2) “Series 0 Channel 0 Green”: 255
3) “Series 0 Channel 0 Blue”: 255
Note: These settings will make the DIC look like it does when you are imaging (no colors added).
DAPI
1) “Series 0 Channel 1 Red”: 0
2) “Series 0 Channel 1 Green”: 0
3) “Series 0 Channel 1 Blue”: 255
Note: These settings will make everything imaged in the DAPI channel blue in color.
GFP
1) “Series 0 Channel 2 Red”: 0
2) “Series 0 Channel 2 Green”: 255
3) “Series 0 Channel 2 Blue”: 0
Note: These settings will make everything imaged in the GFP/AlexaFluor488 channel green in color.
Press “OK”. Now the image file will open with a slider to move from one channel to another.
Open the “ROI Manager” window: Edit – Selection – Add to Manager.
Open the “Results” window: Analyze – Measure.
Make sure the “Results” window has the following measurements: “Area”, “Mean”, and “IntDen”.
If these are not shown in the “Results” window, right click the gray bar in the “Results” window and select “Set Measurements…”.
Select “Area”, “Integrated density”, and “Mean gray value”. Press “OK”.
Move to the NR fat staining channel. Select the oval tool and draw a small shape in an area with just the background in it (no worms). Measure the background: Analyze – Measure. Background measurement will show up in the “Results” window. This background measurement is used to adjust the fluorescence of the worm, to account for any autofluorescence in the image.
Note: You need a new background measurement for each image analyzed. If you have multiple worms in one image, you can use the same background measurement for all of them. You can use this oval measurement again if you save it in the “ROI Manager” window.
While the oval is still drawn on the image, select the “Add [t]” option in the “ROI Manager” window.
Rename to “Background” to remember to use for future images: Highlight number, select “Rename…”, rename the image and press “OK”.
Move to the DIC channel. Select the polygon tool. Outline the worm completely.
Note: In order to include a worm for analysis, the whole worm must be present in the image.
With the worm still outlined, move to the NR fat staining channel. Add the outline to the “ROI Manager” (“Add [t]”). Measure the fluorescence in the outlined worm (Analyze – Measure).
Close image: File – Close.
Repeat for all other images in the analysis.
Open image: File – Open.
Move to the NR fat staining channel.
Background measurement: Select a ROI in the “ROI Manager” labelled background. Measure background: Analyze – Measure.
Worm measurement: Move to the DIC channel, select the polygon tool, and outline the worm. Move to the NR fat staining channel, add to the “ROI Manager” (“Add [t]”). Measure the worm fluorescence: Analyze – Measure.
Close image: File – Close.
Once finished with a strain/condition, save the ROIs in the “ROI Manager”: Highlight all of the ROIs and select: “More…” then “Save” and save.
Save all the measurements by selecting all measurements and copying to a spreadsheet where the area-corrected worm fluorescent intensity (CTCF) will be calculated.
To calculate the CTCF for each image: Multiply the background area by the background mean. Subtract this value from the worm integrated densitiy. Divide by the worm area. The final value is the CTCF.
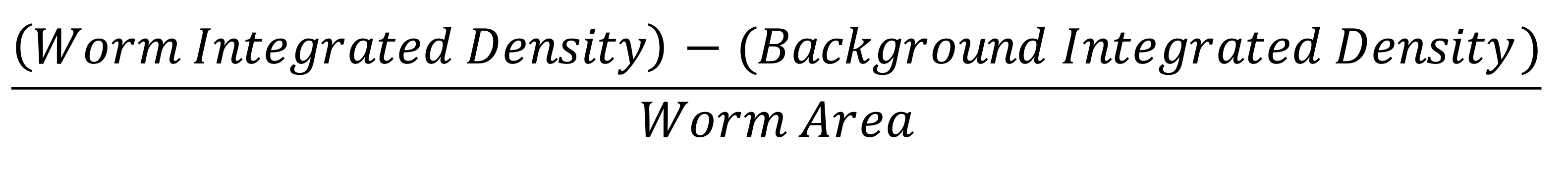
To normalize all samples to the control, calculate the average CTCF value for the control samples. Divide all CTCF values (including the controls) by the average control CTCF. Now everything is normalized to the control.
Plot CTCF values on a graph. If there are multiple samples, either a t-test or ANOVA can be performed to determine significant differences between the samples’ fat content. Representative quantification of data is shown in Figure 4.
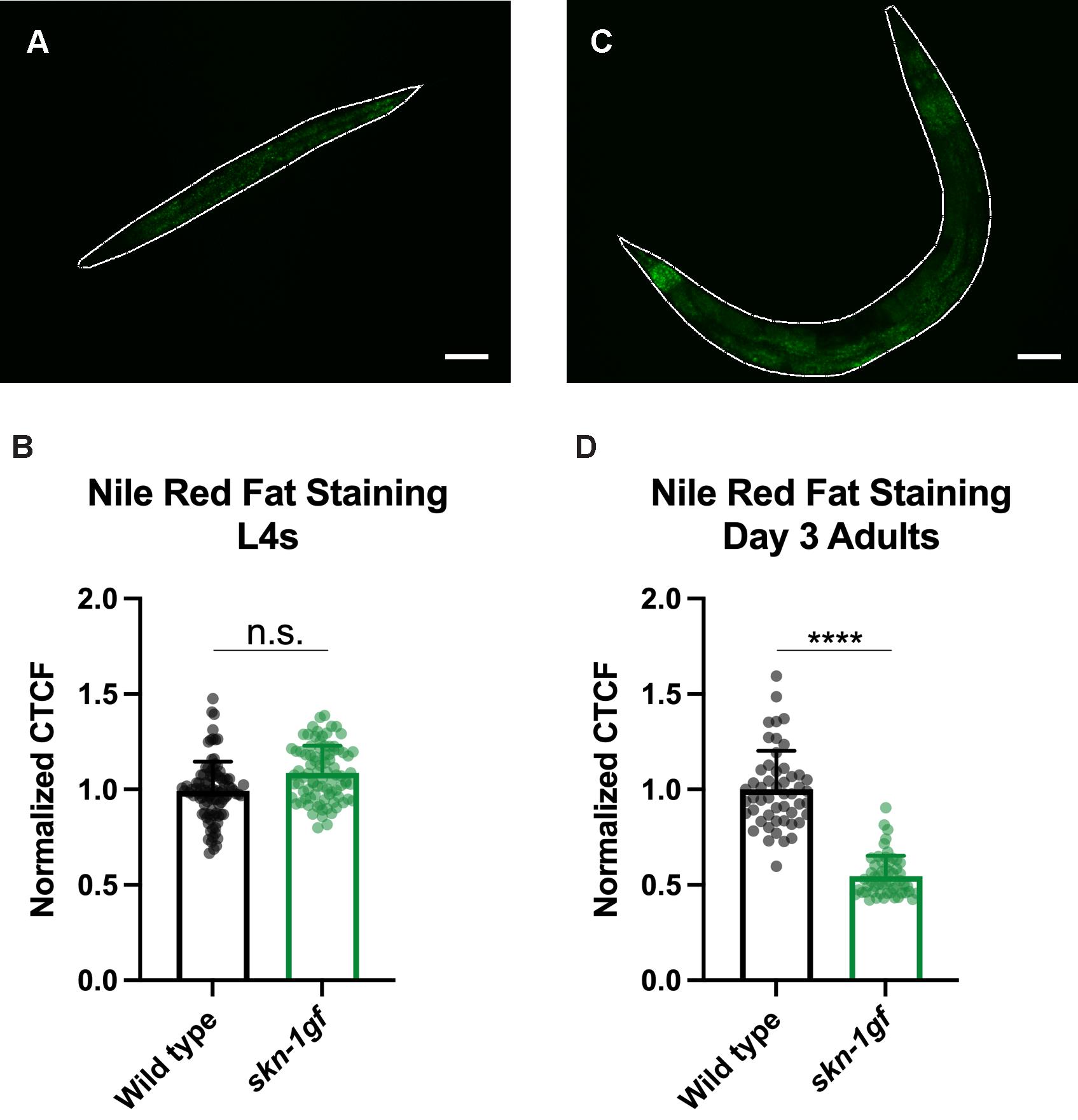
Figure 4. Quantification of C. elegans stained with Nile Red. One worm has been outlined (in white) in ImageJ for size analysis and lipid density (total fluorescence). Scale bar = 50 μm. Quantification of lipid abundance in populations of animals of the indicated genotypes [n = 100 for (B) and n = 50 for (D)]; representative images found in Figure 2).
Quantitative analysis of ORO-stained animals
ORO-stained animals are used to measure fat droplet sizes and fat distribution throughout the body. For example, ORO can be used to determine the category of fat distribution in animals that have undergone Asdf as previously described (Nhan et al., 2019).
Notes
NGM stock plates should be freshly prepared and freshly seeded for all experiments.
Staining solutions should be prepared on the day of staining (NR) or the day before staining (ORO).
Always include a control group to make comparisons within each experimental group.
Although the washing steps use PBS + 0.01% Triton X-100, worms and embryos tend to stick to the side of the tubes. It is recommended to have over double the population required for imaging, to compensate for the loss of some of the sample.
Recipes
Nematode Growth Media (NGM) plates
Prepare stock solutions of 1 M MgSO4, 5 mg/mL cholesterol, 1 M KH2PO4, 1 M CaCl2, and 2.5% (w/v) streptomycin.
Dissolve 120.366 g of MgSO4 per 1 L of water (1 M). Filter sterilize.
Dissolve 5 g of cholesterol per 1 L of 100% ethanol (5 mg/mL). Store at 4°C.
Dissolve 136.086 g of KH2PO4 per 1 L of water (1 M). Filter sterilize.
Dissolve 110.98 g of CaCl2 per 1 L of water (1 M). Filter sterilize.
Dissolve 25 g of streptomycin sulfate per 1 L of water (2.5%). Filter sterilize. Store at 4°C.
Add 3 g of NaCl, 17 g of agar, 2.5 g of peptone, and 950 mL of water to a glass flask. Include a stir bar.
Cover with foil and autoclave to sterilize and dissolve agar.
Cool, with stirring, to 55–60°C.
Add 1 mL of 1 M MgSO4, 1 mL of 5 mg/mL cholesterol, 25 mL of 1 M KH2PO4, 1 mL of 1 M CaCl2, and 7.5 mL of 2.5% streptomycin.
Stir for 15 min.
Dispense 11 mL into 6 cm Petri dishes, using a sterile technique.
Allow to solidify overnight.
Seed stock NGM plates with 250 μL of OP50-1 overnight culture (grown in LB + streptomycin, without shaking).
Allow plates to dry (covered) for 2–3 days before use.
Phosphate buffered saline (PBS)
Add 26.5 g of Na2HPO4·H2O, 80 g of NaCl, 2 g of KCl, and 2 g of KH2PO4 to a glass bottle.
Bring volume to 1 L with water.
Sterilize by autoclaving.
PBS + 0.01% Triton X-100 (PBST)
Using a cut pipette tip (to broaden the opening), add 100 μL of Triton X-100 to 1 L of sterile PBS.
Mix well (do not shake).
M9
Add 30 g of KH2PO4, 60 g of Na2HPO4, 50 g of NaCl, and 120 mg of MgSO4 to a glass bottle.
Bring volume to 1 L with water.
Sterilize by autoclaving.
Precipitation of calcium salts may occur after autoclaving. Re-dissolving may take several days. Agitation helps.
DAPI stock solution
Dissolve 1 mg/mL in DMSO.
ORO stock solution
Add 1.5 g ORO to 300 mL of 100% isopropanol in a glass bottle for a final concentration of 5 mg/mL. Add a stir bar.
Let stir overnight. Store at room temperature in the dark covered in foil (can be stored for months).
Optional: Filter the stock solution after stirring, before storing in the dark at room temperature.
NR stock solution
Cover a glass bottle with foil.
Add 100 mg NR for microscopy to 200 mL of 100% acetone in a glass bottle. Add a stir bar.
Let stir overnight in the dark. Store at room temperature in a dark place covered in foil (can be stored for months).
Acknowledgments
Protocol is derived from the original research paper, Nhan et al. “Redirection of SKN-1 abates the negative metabolic outcomes of a perceived pathogen infection” Proc Natl Acad Sci U S A. 2019 Oct 29;116(44):22322-22330. doi: 10.1073/pnas.1909666116 (Nhan et al., 2019). This work was funded by the NIH R01AG058610 to S.P.C., T32AG052374 to N.L.S and B.V.C., T32GM118289 to N.L.S., and T32AG000037 to J.D.N. and A.M.H.
Competing interests
The authors declare no competing interests.
Ethics
No human or vertebrate animal subjects are used in this study.
References
- Castro, C., Sar, F., Shaw, W.R., Mishima, M., Miska, E. A. and Griffin., J. L. (2012). A metabolomic strategy defines the regulation of lipid content and global metabolism by Delta9 desaturases in Caenorhabditis elegans. BMC Genomics 13: 36.
- Greenspan, P., Mayer, E. P. and Fowler., S. D. (1985). Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100(3): 965-973.
- Hammerquist, A. M., Escorcia, W. and Curran, S. P. (2021). Maf1 regulates intracellular lipid homeostasis in response to DNA damage response activation. Mol Biol Cell 32(11): 1086-1093.
- Johnson, A. A. and Stolzing, A. (2019). The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell e13048.
- Lynn, D. A., Dalton, H. M., Sowa, J. N., Wang, M. C., Soukas, A. A. and Curran, S. P. (2015). Omega-3 and-6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc Natl Acad Sci U S A 112(50): 15378-15383.
- Nhan, J. D., Turner, C. D., Anderson, S. M., Yen, C. A., Dalton, H. M., Cheesman, H. K., Ruter, D. L., Uma Naresh, N., Haynes, C. M. and Soukas, A. A. (2019). Redirection of SKN-1 abates the negative metabolic outcomes of a perceived pathogen infection. Proc Natl Acad Sci U S A 116(44): 22322-22330.
- O'Rourke, E. J., Soukas, A. A., Carr, C. E. and Ruvkun, G. (2009). C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metabolism 10(5): 430-435.
- Pang, S. and Curran, S. P. (2014). Adaptive Capacity to Bacterial Diet Modulates Aging in C. elegans. Cell Metabolism 19(2): 221-231.
- Pino, E. C., Webster, C. M., Carr, C. E. and Soukas, A. A. (2013). Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J Vis Exp JoVE(73).
- Stuhr, N. L. and Curran, S. P. (2020). Bacterial diets differentially alter lifespan and healthspan trajectories in C. elegans. Commun Biol 3(1): 653.
- Webster, C. M., Pino, E. C., Carr, C. E., Wu, L., Zhou, B., Cedillo, L., Kacergis, M. C., Curran, S. P. and Soukas, A. A. (2017). Genome-wide RNAi screen for fat regulatory genes in C. elegans identifies a proteostasis-AMPK axis critical for starvation survival. Cell Rep 20(3): 627-640.
- Zhang, Y., Zou, X., Ding, Y., Wang, H., Wu, X. and Liang, B. (2013). Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC Genomics 14: 164.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Stuhr, N. L., Nhan, J. D., Hammerquist, A. M., Van Camp, B., Reoyo, D. and Curran, S. P. (2022). Rapid Lipid Quantification in Caenorhabditis elegans by Oil Red O and Nile Red Staining. Bio-protocol 12(5): e4340. DOI: 10.21769/BioProtoc.4340.
Category
Biochemistry > Lipid > Lipid measurement
Cell Biology > Cell staining > Lipid
Cell Biology > Cell metabolism > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









